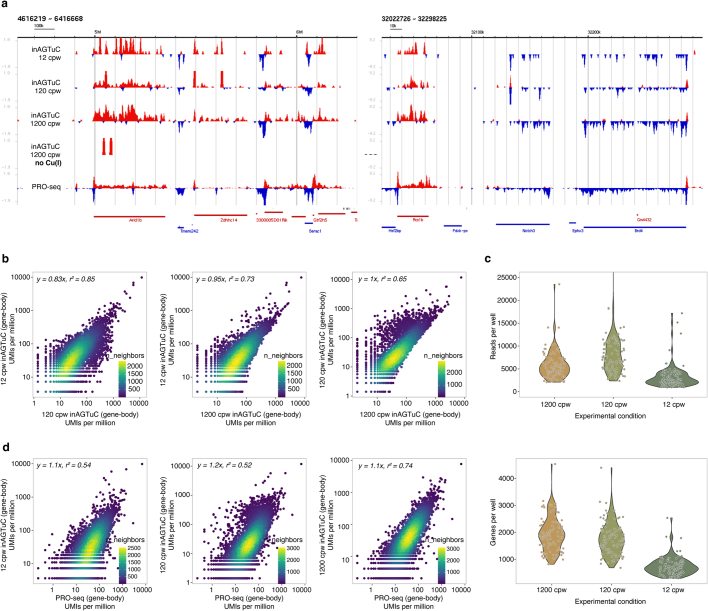
Extended Data Fig. 5. Feasibility demonstration of inAGTuC with fewer cells.
a, Representative genome-browser screenshots of inAGTuC library from one 96-well plate with each well containing either 12, 120, or 1200 cells per well (cpw) showing two regions in chromosome 17 of the mouse genome (mm10). inAGTuC library with 1200 cpw but without Cu(I) in the CuAAC reaction (fourth track) and the PRO-seq library (fifth track) serve as the negative and positive control, respectively. b, Correlations among 12 cpw, 120 cpw, and 1200 cpw inAGTuC libraries in the body of genes (n = 19,961). c, Distribution of UMIs per well (top) and genes per well (bottom) in 12 cpw, 120 cpw, and 1200 cpw inAGTuC libraries. d, Correlations between PRO-seq and 12 cpw, 120 cpw, and 1200 cpw inAGTuC libraries in the body of genes (n = 19,961). For panels b and d, UMIs from the 500 bp regions from each end of the genes were removed to only include nascent RNA from elongating RNA polymerases, and the data was plotted on a log-log scale to show the range of data distribution.