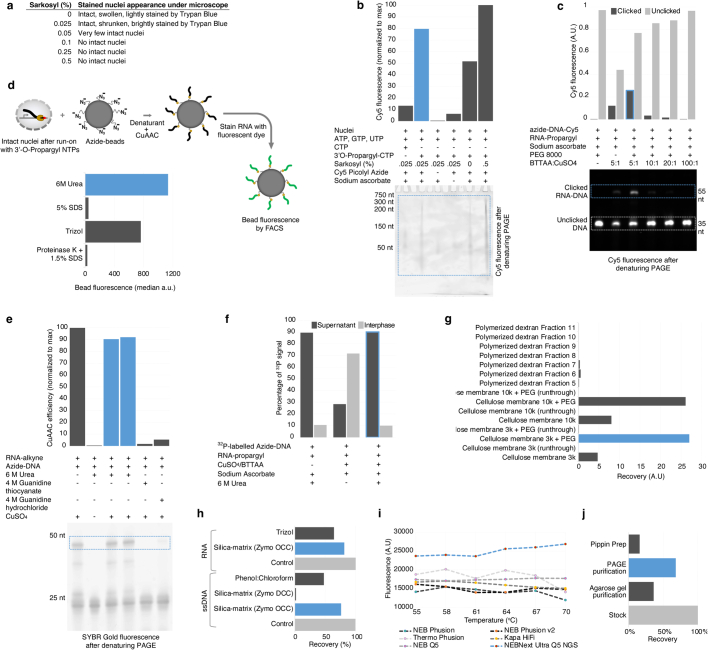
Extended Data Fig. 3. Optimization of intact nuclei run-on reaction and NGS library preparation steps.
a, Physical appearance of Trypan Blue stained nuclei under microscope treated with various sarkosyl concentrations. b, Relative quantification of nuclear run-on efficiency with various sarkosyl concentrations. Nascent RNA collected after nuclear run-on reaction with either native CTP or click-compatible 3′-(O-Propargyl)-CTP was clicked with Cy5-azide, resolved in denaturing PAGE, and imaged for Cy5 fluorescence. c, Effect of different ratios of CuAAC accelerating ligand BTTAA in CuAAC efficiency. RNA-propargyl was clicked with azide-DNA containing Cy5 in the presence of various ratios of BTTAA:CuSO4, resolved in denaturing PAGE, and imaged for Cy5 fluorescence. d, Relative quantification of denaturing efficiency of commonly used denaturing agents to release the nascent RNA from RNA polymerase complex. Intact nuclei after run-on with 3′-(O-Propargyl)-NTPs were treated with denaturing agents in the presence of azide-labeled beads and CuAAC reagents, allowing nascent RNA to click with the beads. Beads were stained with RNA-binding dye and measured for fluorescence by FACS. e, Effect of denaturing agent’s presence in CuAAC efficiency. The blue outline in the image of denaturing PAGE denotes the click product between the RNA-alkyne and azide-DNA. f, Role of urea in the residence of clicked RNA-DNA conjugate in either supernatant or interphase of Trizol during the clean-up of CuAAC reaction, as quantified by the scintillation count of 32P radioisotope. g, Desalting (removal of CuSO4, BTTAA, and sodium ascorbate from CuAAC reaction) efficiency of polymerized dextran and cellulose membrane. Fluorescence from Cy5-labeled RNA-DNA conjugate was measured in elution fractions from columns packed with polymerized dextran and elution from different pore-size cellulose membrane centrifugation tubes with or without PEG 8000. h, Relative recovery of ssDNA or RNA from phenol:chloroform or silica-based matrix column purification. Clicked RNA-DNA conjugate was radioisotope labeled using Polynucleotide kinase and γ-32P ATP, and the cleaned reaction was quantified using a scintillation counter. i, PCR amplification efficiency of clicked RNA-DNA conjugate using different commercial PCR amplification kits. The PCR reaction was resolved in native PAGE, stained with SYBR Gold, and quantified using ImageJ software. j, Relative recovery of size-selected dsDNA. A mock NGS library (purified PCR product) was selected for the desired size using various size-selection methods, and the recovered dsDNA was quantified using a dsDNA-specific fluorescence kit (Qubit). The bar represents the average of two independent replicates. Note: The blue bar, line, or border represents the “winner” condition. Polyacrylamide gel electrophoresis for b, c, and e was repeated at least twice with the addition or subtraction of some conditions presented here. For gel source data, see Supplementary Fig. 2.