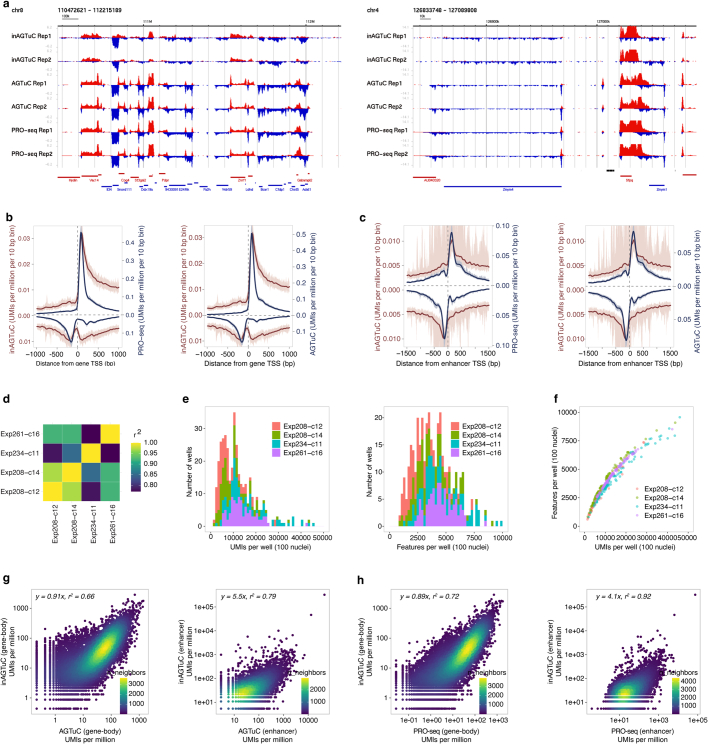
Extended Data Fig. 4. Benchmarking inAGTuc against AGTuC and PRO-seq.
a, Representative genome-browser screenshots with two replicates of inAGTuC, AGTuC, and PRO-seq showing a region in chromosome 8 (left) and a region in chromosome 4 of the mouse genome (mm10). b, c, Comparison of inAGTuC metagene profiles with PRO-seq and AGTuC using UMIs per million per 10 base pair bins around (b) the TSS of genes (n = 19,961) and (c) enhancers (n = 12,542). The line represents the mean, and the shaded region represents a 95% confidence interval. d, Correlations of inAGTuC UMIs per million sequences in gene bodies (n = 19,961) between the four replicates. e, Distribution of UMIs per well (left) and features per well (right) in four replicates of 96-well plate inAGTuC libraries. Each well contains 100 nuclei. f, Relationship between the UMIs per well and the number of features detected per well in four replicates of 96-well plate inAGTuC libraries. g, Correlation between inAGTuC and AGTuC UMIs per million sequences in the body of genes (n = 19,961) and enhancers (n = 12,542). h, Correlation between inAGTuC and PRO-seq UMIs per million sequences in the body of genes (n = 19,961) and enhancers (n = 12,542). For panels g and h, UMIs from the 500 bp regions from each end of the genes and 250 bp regions from each end of the enhancers were removed to only include nascent RNA from elongating RNA polymerases, and the data was plotted on a log-log scale to show the range of data distribution.