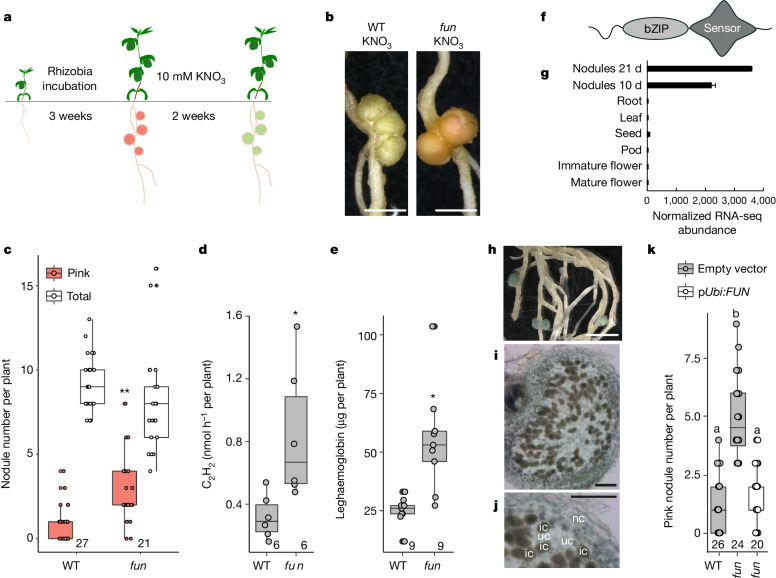
Fig. 1. FUN is essential for the suppression of nitrogen fixation by environmental nitrate.
a, Schematic diagram of the screen that resulted in identification of FUN. Mutants producing functionally pink nodules were watered with 10 mM KNO3. Most nodules on wild-type plants became green and senescent, whereas fun mutant plants maintained pink nodules even under high concentrations of nitrate. b–e, Nodulation phenotypes of fun mutants in high-nitrate conditions. The nodule appearance (b), nodule number (c), nitrogen fixation measured by acetylene reduction assay (ARA) (d) and leghaemoglobin content (e) of fun mutant plants after 2 weeks of exposure to 10 mM KNO3. b, Scale bars, 1 cm. f, Schematic of the FUN protein, showing bZIP DNA-binding and sensor domains. g, The expression pattern of FUN in different tissues obtained from the Lotus expression atlas42. h–j, The expression pattern of the FUN promoter is revealed by beta-glucuronidase (GUS) reporter gene expression. FUN is expressed exclusively in nodules (h,i), and cross-sections (i,j) indicate that FUN is expressed predominantly in uninfected cells (uc) and nodule cortex (nc), and to a lesser extent in infected cells (ic) (j). Scale bars: 2 cm (h), 200 µm (i,j). k, Complementation of fun mutants using expression of FUN–GFP under the control of the Lotus ubiquitin promoter restores the sensitivity of fun nodules to nitrate. Letters indicate groups that are significantly different from each other (P < 0.05). c–e,k, In box plots, the centre line represents the median, box edges delineate first and third quartiles, whiskers extend to maximum and minimum values and dots show individual values. Numbers below data in box plots represent the number of biologically independent samples. P values determined by ANOVA and Tukey post hoc testing; *P < 0.05, **P < 0.01.