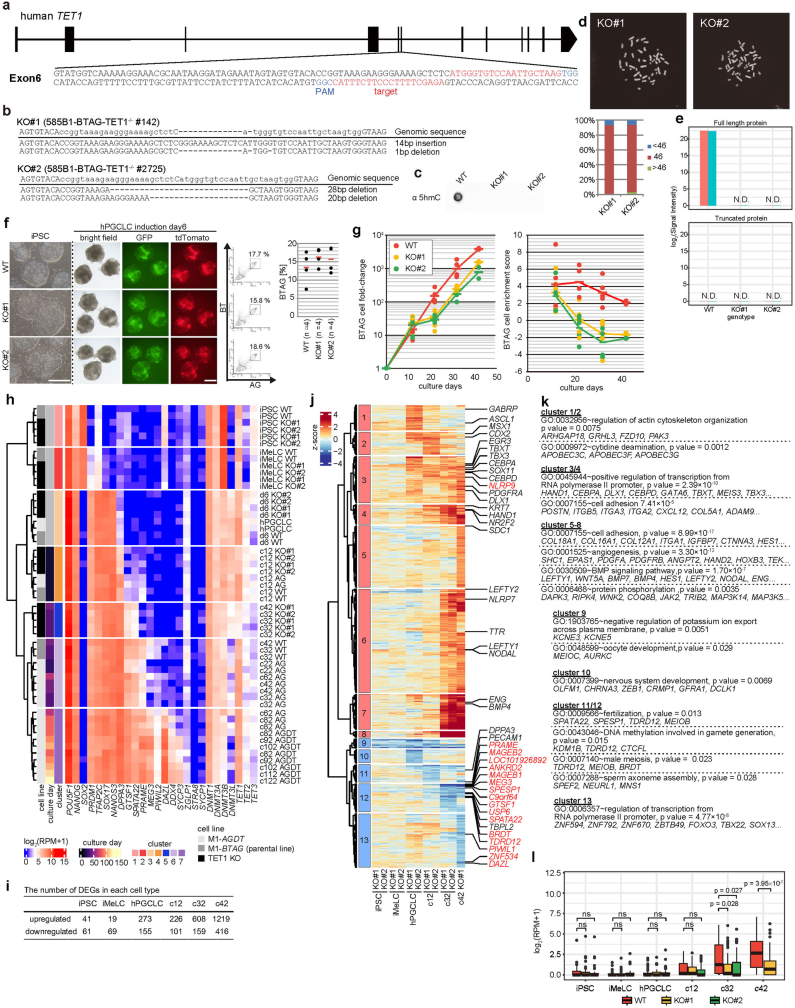
Extended Data Fig. 10. Generation of TET1 knockout hiPSCs and analysis of BMP-driven TET1-knockout hPGCLC differentiation.
a, Scheme of the human TET1 locus, with the illustration of PAM (protospacer adjacent motif) and guide RNA sequences in the exon 6. Black boxes indicate the exons. b, Sequences of the targeted loci in two TET1 knockout (KO) cell lines [TET1 KO#1 and # 2 (M1-BTAG TET1−/− #142 and #2725)]. c, Dot-blot analysis of genomic 5hmC levels in wild-type and TET1 KO hiPSCs (1 replicate for each line). d, Karyotype of TET1 KO#1 and #2 hiPSCs (top: chromosome spreads; bottom: percentage of cells with 46 or other chromosome numbers) (1 biological replicate for each line). Bar, 10 μm. e, Mass spectrometric analysis [log2(signal intensities)] for TET1 and its truncated protein potentially derived from the TET1 KO allele in wild-type and TET1 KO cells. Peptides from the full-length (top), but not the truncated (bottom), TET1 were detected from the wild-type cells (red and blue bars), whereas neither form was detected from the TET1 KO cells (2 biological replicates). f, Induction of hPGCLCs from wild-type (M1-BTAG) and TET1 KO#1 and #2 hiPSCs. Photomicrographs of hiPSCs and iMeLC aggregates induced for hPGCLCs for 6 days (bright-field and fluorescence images for AG and BT) (left), their flow cytometric plots for AG and BT expression (middle), and percentages of BT+AG+ cells (right) from each genotype are shown (4 biological replicates). Bar, 500 μm. g, Growth curves of BT+AG+ cells and enrichment scores during BMP-driven (~c12: 25 ng/ml; c12~: 100 ng/ml) wild-type and TET1 KO#1 and #2 hPGCLC differentiation. 5 and 2 biological replicates for c12−c32 and for c42, respectively. The color coding is as indicated. h, UHC of the transcriptomes during hPGCLC induction and BMP-driven hPGCLC differentiation from wild-type and TET1 KO hiPSCs, with the expression levels of key genes indicated. The color coding is as indicated. i−k, The numbers of the differentially expressed genes (DEGs) [log2(RPM + 1) ≥ 3, fold change ≥ 2] between wild-type and TET1 KO cells (up- or down-regulated in TET1 KO cells) (i), UHC of the DEGs (j), and the GO enrichments and representative genes in the indicated DEG clusters (k). DEGs were defined using average expression values of biological replicates. The DEG numbers were unions of two comparisons (i.e., wild-type vs KO#1 and vs KO#2). Core ER genes were highlighted in red in (j). l, Box plots for the expression dynamics of ER genes (n = 42) during hPGCLC induction and BMP-driven hPGCLC differentiation from wild-type and TET1 KO hiPSCs. p-values of Two-sided Dunnet’s test (except c42) or paired two-sided t-test (c42) were shown. The upper hinges, lower hinges, and middle lines indicate 75 percentiles, 25 percentiles, and median values, respectively. The whiskers are drawn in length equal to the inter-quartile range (IQR) multiplied by 1.5. Data beyond the upper/lower whiskers are not shown.