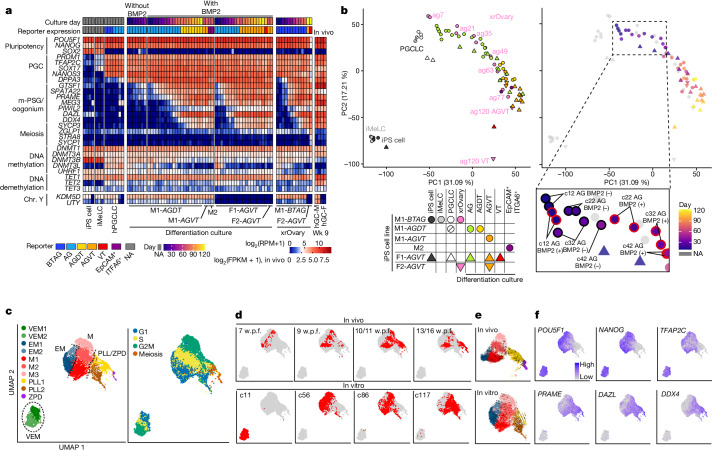
Fig. 2. Transcriptome dynamics during BMP-driven hPGCLC differentiation.
a, Heatmap showing the expression levels of the indicated genes in the indicated cell types (see Supplementary Table 2 for full sample information). Colour coding is as indicated. FPKM, fragments per kilobase million; NA, not applicable; RPM, reads per million. b, PCA of transcriptomes during hPGCLC induction and BMP-driven or xrOvary-based hPGCLC differentiation. The left and right panels are colour coded with reporter expression and culture days, respectively, as indicated. The dotted area in the chart on the right is magnified to clarify the difference of transcriptome progression between cultures with BMP2 (BMP2 (+)) and without BMP2 (BMP2 (−)). c, Uniform manifold approximation and projection (UMAP) and Louvain clustering of scRNA-seq data of female germ cells at 7−16 w.p.f. in vivo21,22 and F1-AGVT hPGCLC-derived cells at c11, c56, c86 and c117 in vitro. Cell-type (left) and cell cycle (right) annotations are shown. VEM, very early mitotic; EM, early mitotic; M, mitotic; PLL, preleptotene and leptotene; ZPD, zygotene, pachytene and diplotene. d−f, UMAP plots as in c, with the annotation of in vivo and in vitro samples (d), with potential developmental trajectories of in vivo and in vitro cell types analysed by RNA velocity (e), and with expression levels of the indicated genes (f). Colour coding is as indicated.