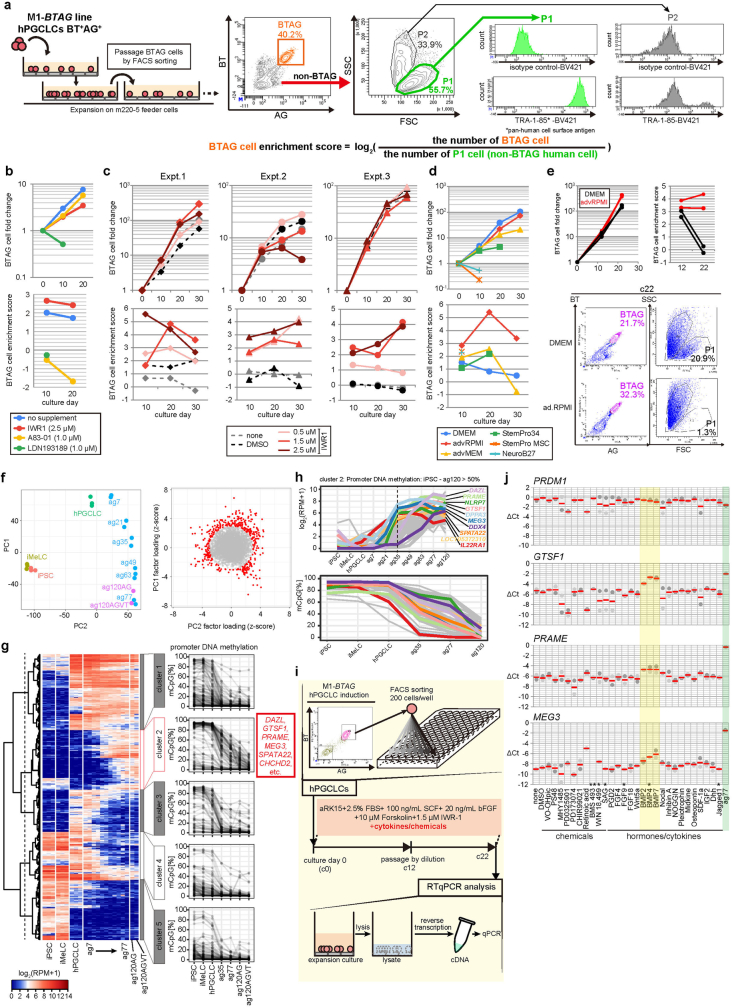
Extended Data Fig. 1. Exploration of the signaling for hPGCLC differentiation.
a, Scheme for hPGCLC expansion culture (left)12 and flow cytometric plot for BTAG expression of the hPGCLC culture and for forward and side scatter (FSC and SSC) of the non-BTAG cells (middle). The P1 cells in the middle panel are TRA-1-85+ (a human-specific antigen)95, i.e., de-differentiated hPGCLC-derived cells, whereas a majority of the P2 cells are TRA-1-85−, i.e., m220 feeders (right). Accordingly, the enrichment score is defined as log2 (the number of BT+AG+ cells/the number of cells in the P1 gate) (right). hPGCLCs were cultured as in12. See Fig. 1a for the summary of acronyms used in this study. b−d, hPGCLC expansion and the enrichment score of the hPGCLC culture with IWR196, A83-01, and LDN193189 at culture day (c) 10 and 20 (b), with different doses of IWR1 at c10, 20, and 30 (c), and with different basal media (d). The passages were performed using flow cytometry. The color coding is as indicated. hPGCLCs were cultured as in12 with or without indicated chemicals. 1 biological replicate for (b) and (d), and 3 biological replicates for (c). e, hPGCLC expansion and the enrichment score of the hPGCLC culture with IWR1 (1.5 μm) in DMEM or advanced RPMI at c12 and 22 (top), and FACS plots for BTAG expression and FSC/SSC of the non-BTAG cells of the hPGCLC culture with IWR1 (1.5 μm) in DMEM or advanced RPMI at c22 (bottom). The passages were performed with dilution. The color coding is as indicated. Note that there were nearly no de-differentiated cells in the P1 gate in the culture with advanced RPMI. The data show (top)/represent (bottom) 2 biological replicates. f, Principal component analysis (PCA) of transcriptomes of key cell types during hPGCLC induction and hPGCLC differentiation in xrOvaries14 (top) and the identification of genes making significant contributions [radius of standard deviations (SDs) ≥ 3] to scaled PC1 and PC2 loadings (bottom). Genes expressed in at least one sample [log2(RPM + 1) ≥ 4] were used for PCA. g, (left) Unsupervised hierarchical clustering (UHC) of the genes selected in (f) based on their expression dynamics, and (right) promoter methylation dynamics of the genes in the five clusters in (left) during hPGCLC induction and hPGCLC differentiation in xrOvaries14. Among the cluster 2 genes, those showing promoter 5mC-level reduction from human iPS cells (hiPSCs) to oogonia-like cells by ≥ 50% are defined as epigenetic reprogramming-activated genes (ER genes). h, Expression (top) and promoter methylation (bottom) dynamics of epigenetic reprogramming-activated genes (ER genes) during hPGCLC differentiation in xrOvaries. Top eight ER genes in the expression level at ag35, and DAZL and DDX4 are annotated. i, Scheme for the screening of cytokines/chemicals that induce ER gene up-regulation. j, Expression of PRDM1, GTSF1, PRAME, and MEG3 measured by qRT-PCR at culture day (c) 22 with the indicated cytokines/chemicals. For each gene, ∆Ct from the average Ct values of two housekeeping genes, RPLP0 and PPIA (set as 0), were calculated and plotted for 2 biological replicates. Mean values are shown as a red bar. *, **: Not detected or ∆Ct <−10 in one or two replicates, respectively. ag77: expression values in hPGCLC-derived cell at ag77 in xrOvaries14.