Extended Data Fig. 10. F1-score for SV inside clusters of different sizes.
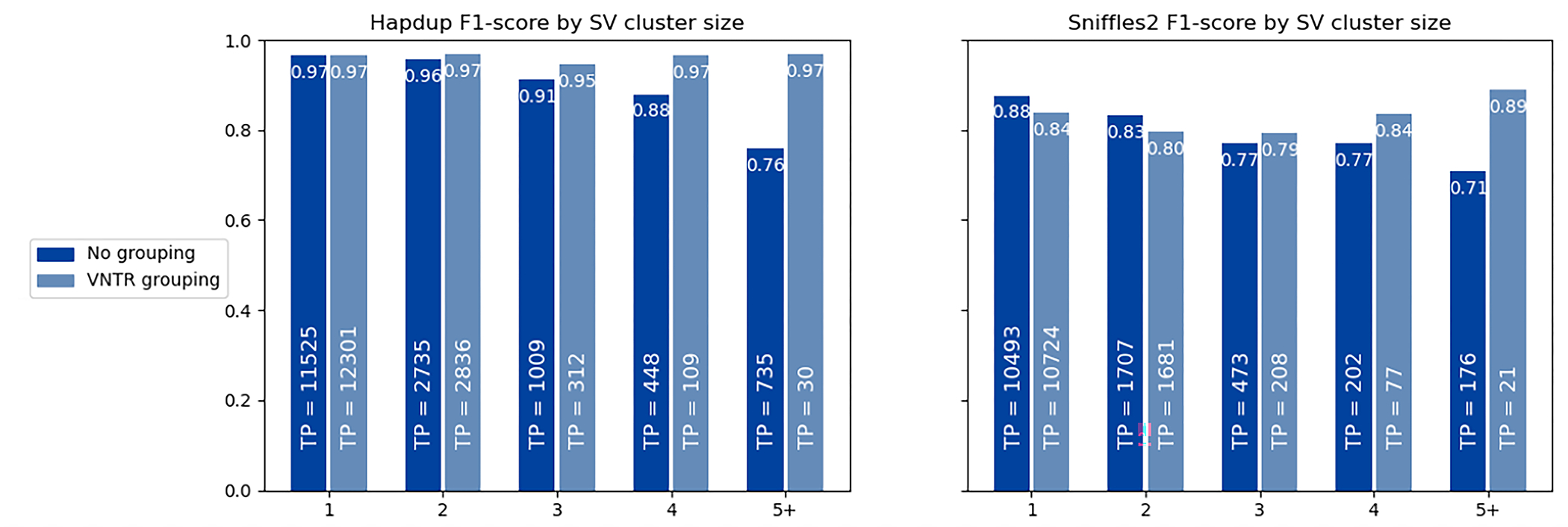
The HiFi calls for HG002 genome were used as reference, and calls within 2 kbp were clustered using single linkage clustering. The number of true positive calls in each category is shown as text. When VNTR grouping is enabled, all insertions and deletions within the same haplotype in a single VNTR are combined into a single call. A substantial portion of the reduced Sniffles2 concordance is explained by the differences in representation of SV clusters by the assembly-based and mapping-based approaches.
