Figure 2. Small variant calling performance evaluation.
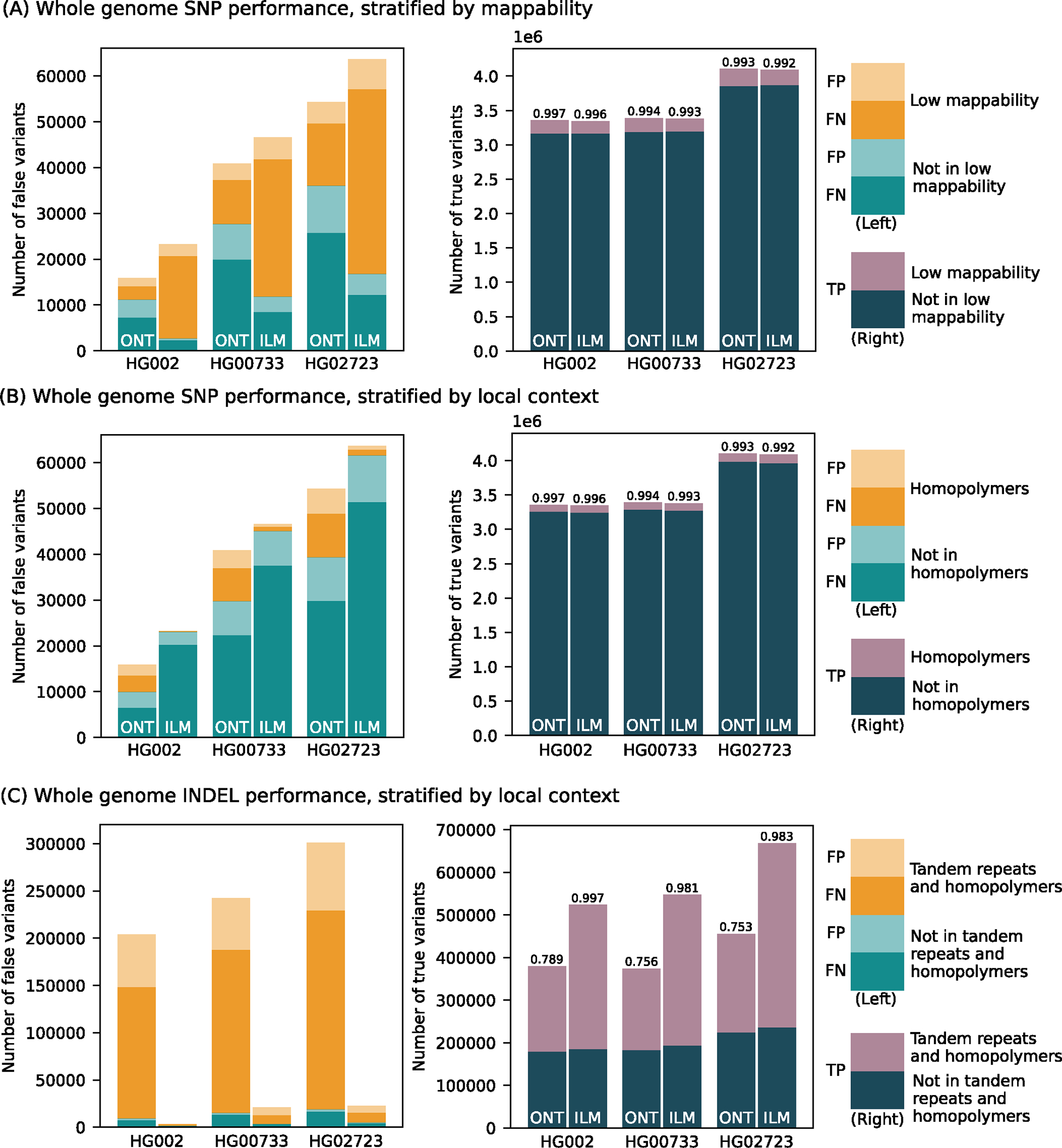
Number of false positive, false negative, and true positive small variant calls made by PEPPER-Margin-DeepVariant (PMDV) using either ONT reads or DeepVariant using Illumina reads. (A) SNP performance, stratified by genomic regions mappability. (B) SNP performance, stratified by homopolymer context. (C) INDEL performance stratified by homopolymers and tandem repeats context. F1-scores are reported on top of the true positive bars. Statistics computed against the Genome in a Bottle v4.2.1 benchmark for HG002; for other cell lines (HG00733, HG02723) small variant calls generated by DeepVariant with HiFi reads are used. Source data is available at Supplementary Table 3.
