Extended Data Fig. 1. Variant calling and methylation analysis using Napu.
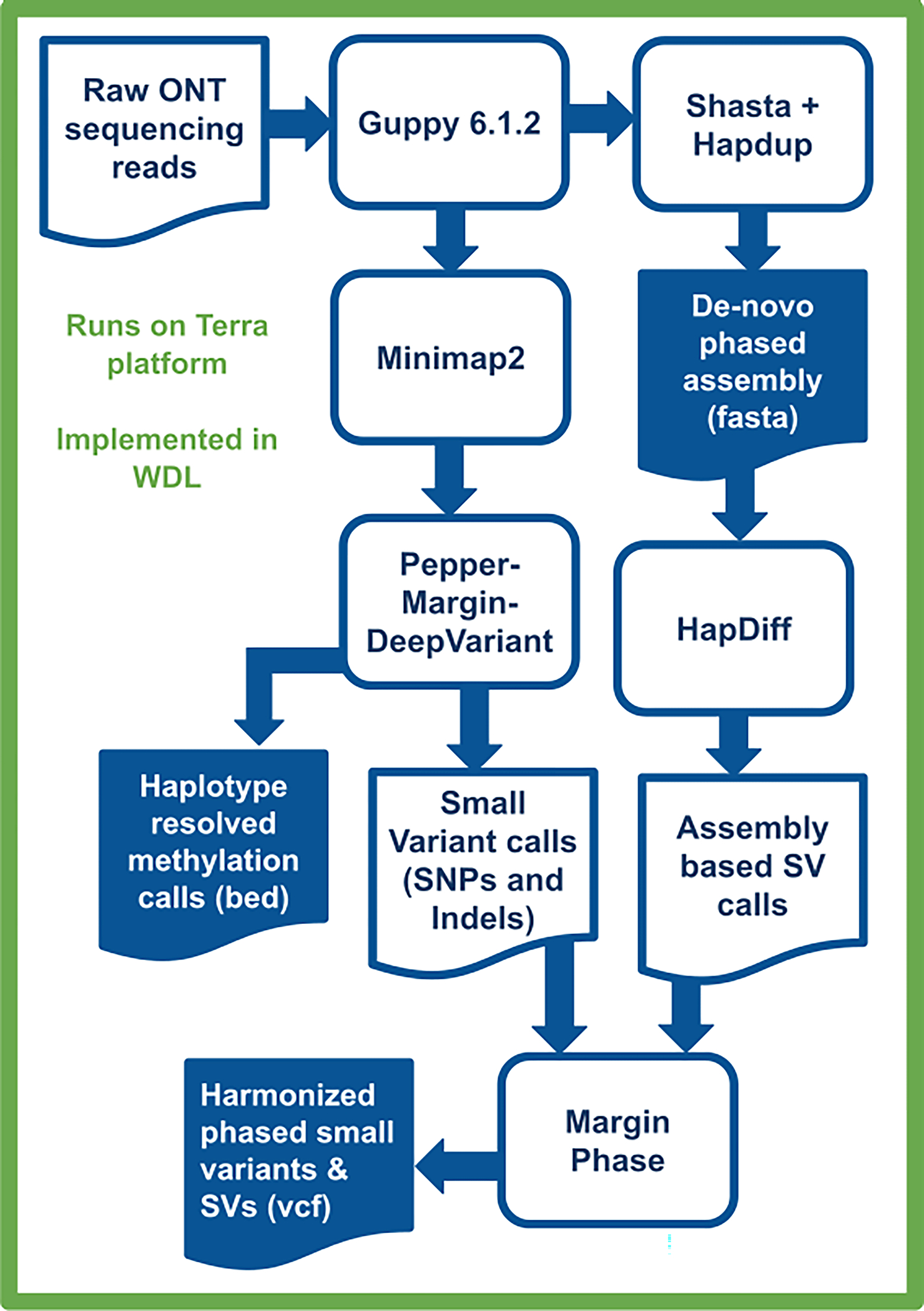
Raw ONT sequencing reads are basecalled by Guppy 6.1.2, which simultaneously produces methylation tags. A diploid, de-novo phased assembly is produced using a combination of Shasta and Hapdup. These assemblies are used to call SVs with Hapdiff. Small variants are called against a reference genome with Pepper-Margin-DeepVariant. The phased alignment file generated by Margin is used to produce haplotype-resolved methylation calls. Small variants and SVs are jointly phased by Margin, producing a single harmonized vcf.
