Extended Data Fig. 3. Assembly metrics comparison against HG002 assemblies produced in Jarvis et. al (2022).
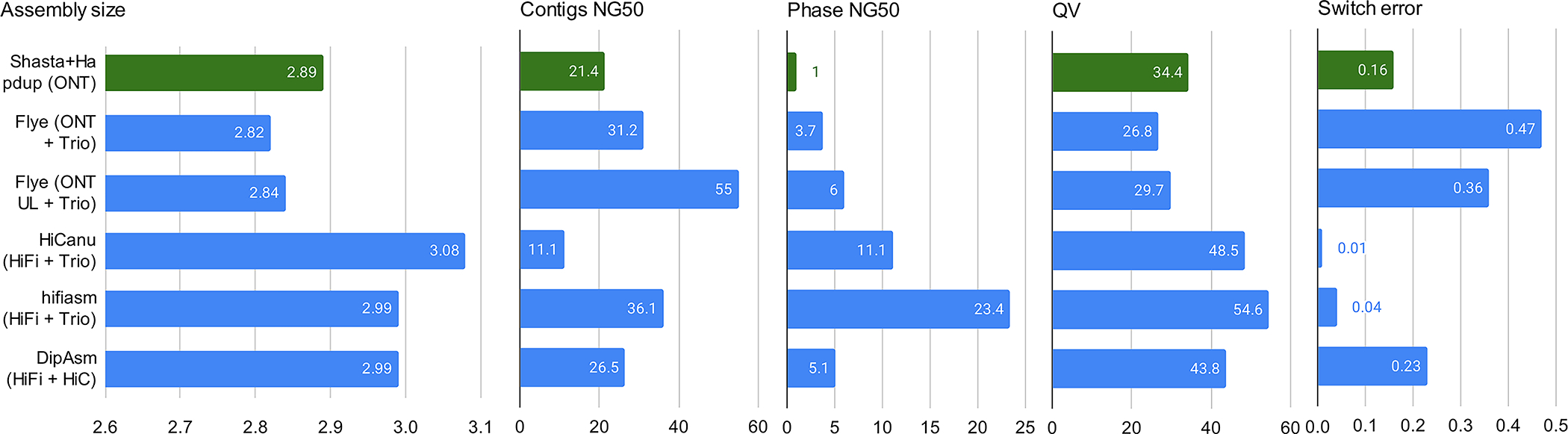
Our assemblies are highlighted in green. Flye (ONT+trio) were produced using standard ONT reads at 60x coverage and Illumina parental information; Flye (ONT UL + trio) is similar, but using ultra-long ONT extraction. HiCanu and hifiasm used 34x HiFi reads and Illumina parental sequencing. DipAsm used 34x HiFi reads and 60x Hi-C reads. Original evaluations from Jarvis et al. are shown. See Supplementary Table 5 for more detail.
