Fig. 2. H3B-8800 treatment induces mis-splicing and down-regulation of DNA damage repair genes.
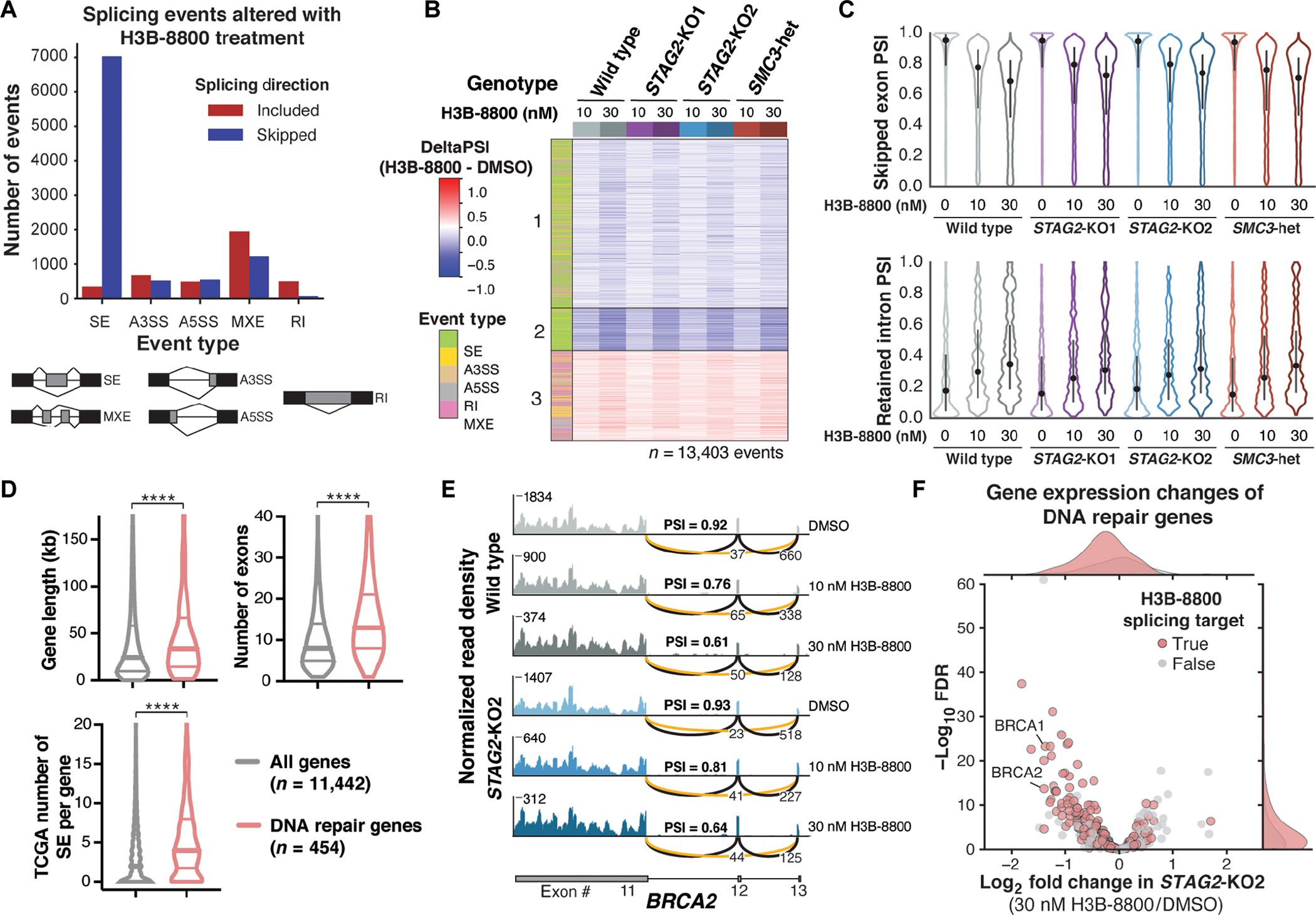
(A) Total number and directionality of splicing alterations induced by 6-hour H3B-8800 treatment in all U937 cell lines. Events are categorized by event type and direction of regulation in H3B-8800–treated relative to DMSO-treated cells. SE, skipped exon; A3SS, alternative 3′ splice site; A5SS, alternative 5′ splice site; MXE, mutually exclusive exon; RI, retained intron. (B) Heatmap of ΔPSI scores for H3B-8800–regulated exons [from (A)] across all conditions separated into three k-means clusters. Each comparison consists of two (STAG2-KO1 and SMC3-heterozygous) or three (STAG2-KO2 and wild type) independent single-cell clones for each concentration of drug compared with DMSO controls within the same genotype. Columns are organized by genotype and concentration of H3B-8800. Color bar on the left indicates the type of splicing event that was called. PSI, percent spliced in. (C) Violin plots showing the distribution of PSI scores for H3B-8800–regulated skipped exons (top) and retained introns (bottom) under each treatment condition tested. Dot represents the median, and bars extend from the first to third quartile range. (D) Violin plots depicting the gene length, number of exons, and number of skipped exons in TCGA comparing DNA repair genes (n = 454) with all expressed protein-coding genes (n = 11,442). Pan-cancer TCGA exon skipping events collated from ExonSkipDB (29). Horizontal lines in violin plots depict the median and first and third quartiles. ****P < 0.0001, Mann-Whitney test. (E) RNA-seq–normalized read density and splice junction track of exon skipping in BRCA2 exon12 from one representative replicate of wild-type and STAG2-KO2 cells treated with DMSO and 10 or 30 nM H3B-8800 for 6 hours. Average PSI scores from three biological replicate samples of exon12 are shown. Average number of reads supporting exon skipping (orange line) and exon inclusion (black line) are reported. (F) Volcano plot of gene expression changes in DNA repair genes in STAG2-KO2 cells treated with 30 nM H3B-8800 relative to DMSO-treated controls. Average log2 fold change of three biological replicates of STAG2-KO2 versus wild-type U937 cells is shown. DNA repair genes that contain H3B-8800–regulated splicing changes are highlighted in red.
