Fig. 6. Splicing changes and down-regulation of DNA repair genes are conserved in patients with MDS and AML.
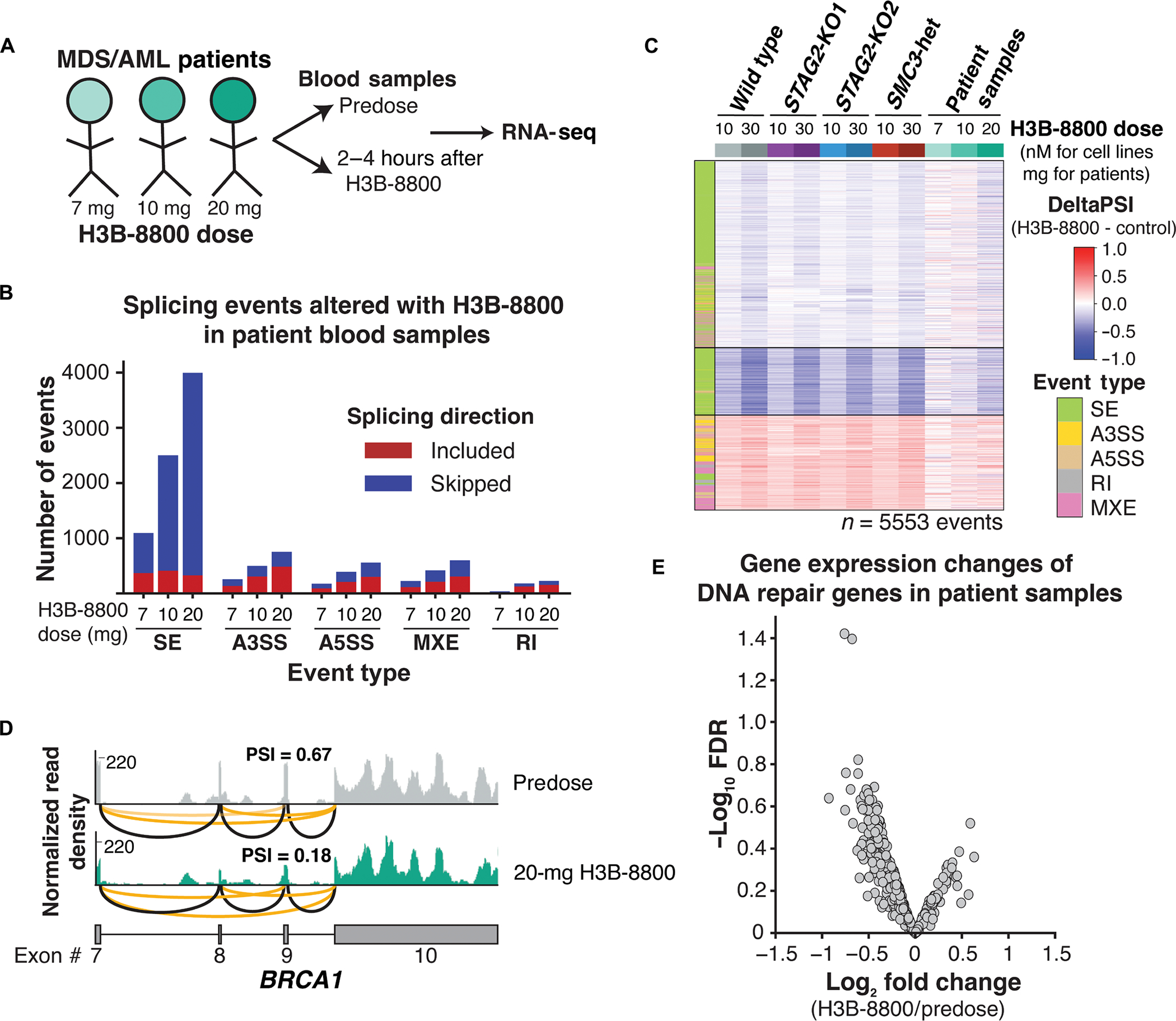
(A) Schematic of samples collected from patients with MDS and AML treated with three different doses of H3B-8800 on clinical trial (clinicalTrials.gov identifier NCT02841540). (B) Total number and directionality of significant (FDR < 0.05, ΔPSI >5%) splicing alterations differentially called in each patient sample pre– and post–H3B-8800. Patients are sorted on the x axis according to increasing doses of H3B-8800. Splicing events are categorized by event type and direction of regulation in H3B-8800 versus pretreatment sample. SE, skipped exon; A3SS, alternative 3′ splice site; A5SS, alternative 5′ splice site; MXE, mutually exclusive exon; RI, retained intron. (C) heatmap of ΔPSI scores for H3B-8800–regulated splicing changes called from U937 cells (Fig. 3a) that are expressed in patient samples. Patient samples are sorted by increasing dose of H3B-8800 received. color bar on the left indicates the type of splicing event that was called, and column colors are labeled by genotype and drug treatment. (D) RNA-seq–normalized read density and splice junction track of exon skipping in BRCA1 exon9 from the pre- and posttreatment sample in the patient who received 20 mg of H3B-8800. Black lines indicate constitutive splicing junctions, and orange lines indicate splice junctions that contain exon skipping. (E) Volcano plot depicting differential gene expression of DNA repair genes from a paired analysis of all patients pre– and post–H3B-8800 treatment.
