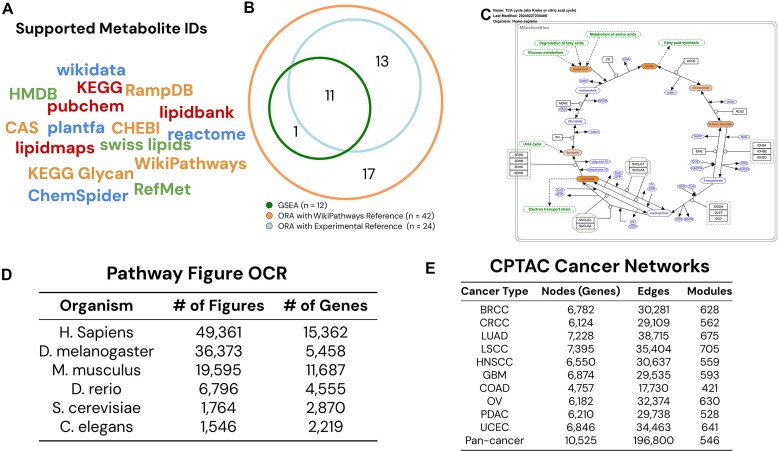
Figure 1.
New data and support for metabolomics. (A) WebGestalt 2024 supports 16 ID types for metabolites with automatic ID detection. (B) Choosing all metabolites in WikiPathways as the reference/background set in ORA results in many enriched pathways not found in GSEA. Limiting the reference set to all experimentally quantified metabolites increases the concordance with GSEA. (C) WebGestalt provides colored pathway maps for WikiPathways and KEGG. For GSEA analysis, identified metabolites are shaded by their input values, as shown in the down-regulated (orange-shade) metabolites in the TCA cycle pathway. (D) WebGestalt 2024 adds tens of thousands of pathways from Pathway Figure OCR for six organisms. (E) WebGestalt 2024 adds 11 new cancer networks generated from CPTAC data and a total of 6488 modules.