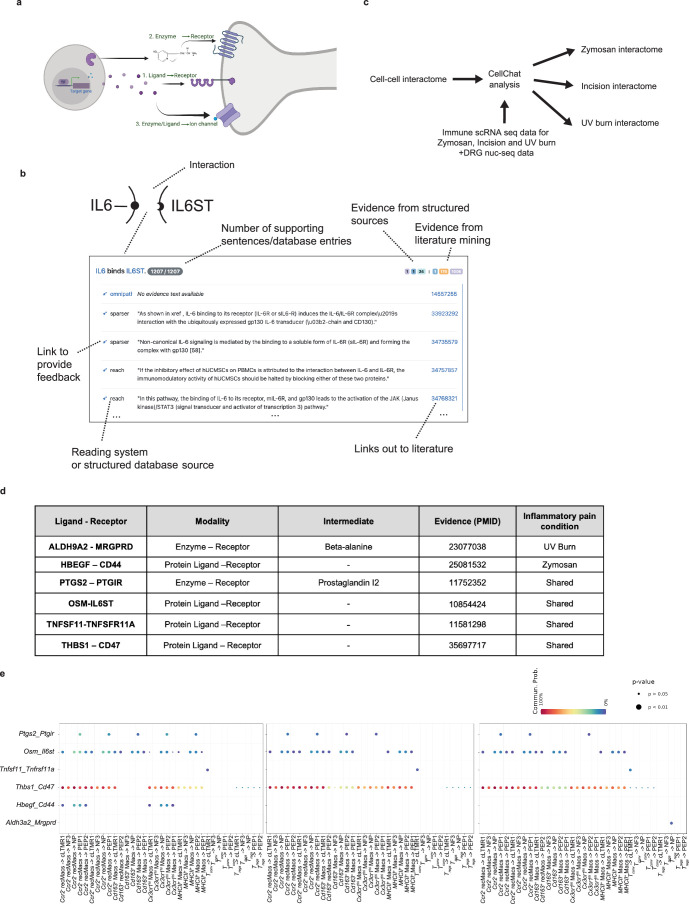
Extended Data Fig. 6. Neuroimmune interactomes of pain have injury-specific signatures.
(a) Schematic showing the types of intercellular communication considered between immune cells and neurons for constructing the interactome. (b) Sample of the web interface for browsing evidence behind the cell-cell interactome. Each interaction is displayed as a heading summarizing the interaction (‘IL6 binds IL6ST’) with gene names linked to HGNC pages representing the gene. The total number of supporting pieces of evidence is shown, as is the breakdown of this number by specific source: different structured databases or literature mining systems integrated with INDRA. Each row under the heading represents a distinct database entry or sentence from a publication, with each publication linked to its corresponding PubMed landing page. Each row also links to a curation page where feedback can be given on the correctness of the interaction. (c) Overview of the condition-specific interactome construction. The general cell-cell interactome was provided as input to Cellchat together with data from immune cells for each of the three pain conditions: UVB, Zymosan, or incision, and combined with naïve DRG neuron data. This results in three condition-specific neuroimmune interactomes. (d) The modalities of interactions and the example of INDRA-based literature evidence, as shown by PMIDs for the representative interactions, are shown in Fig. 4d. (e) Dot plots showing examples of interactions between immune cells and DRG neurons that are shared or injury-specific.