Fig. 2.
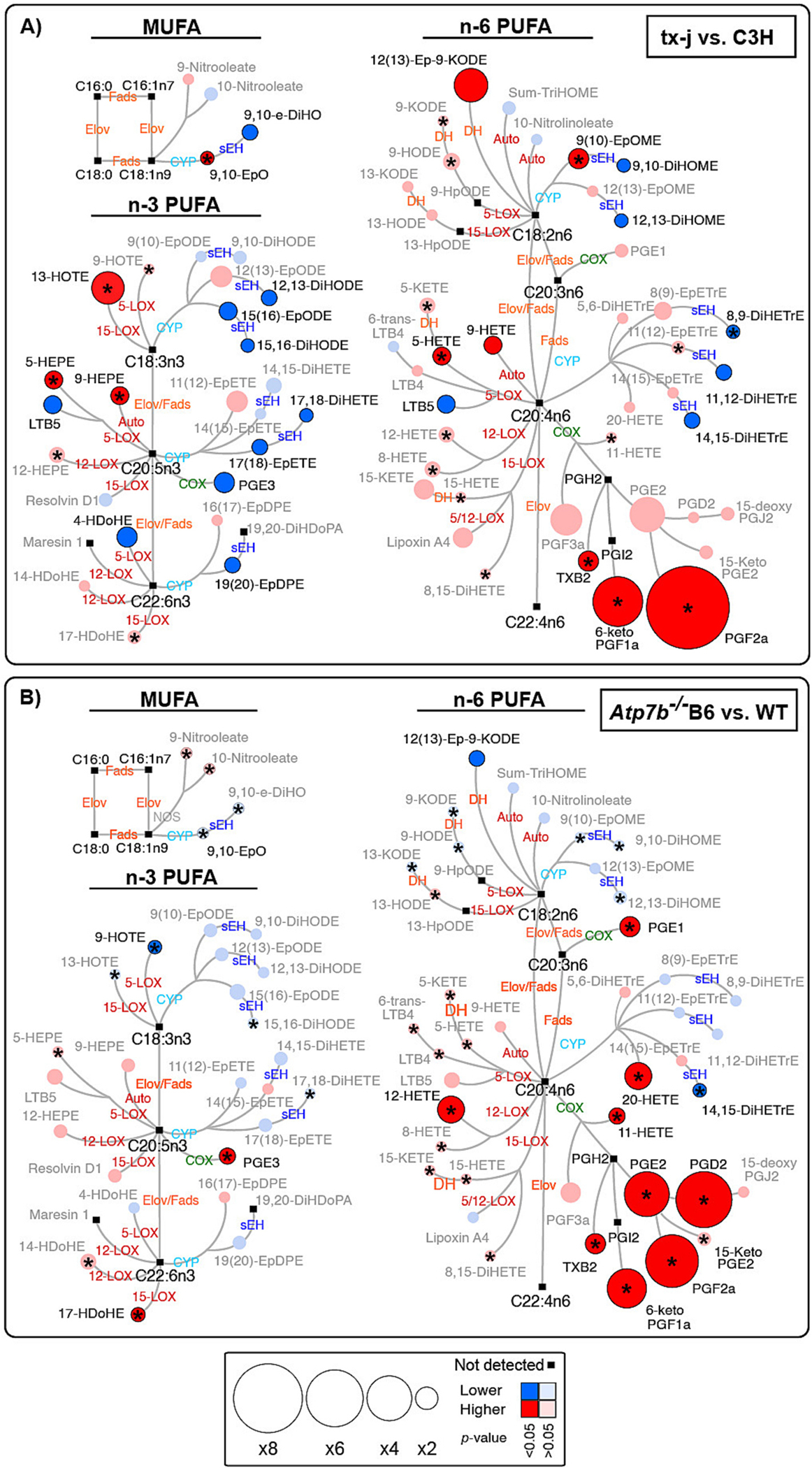
Differences in hepatic oxylipin profiles in two disease mouse models of Wilson’s disease compared with controls. Metabolic network for (A) tx-j vs. C3H and (B) Atp7b−/− B6 vs. WT illustrating metabolic pathways for monounsaturated (MUFAs), n-3 and n-6 polyunsaturated fatty acids (PUFAs), and for oxylipin biosynthesis. MUFAs and PUFAs are described by number of carbons and double bonds (i.e. C18:2n6). Nodes size represents fold changes, calculated as A/B where A is the geometric mean of (tx-j or Atp7b−/− B6) and B is the geometric mean of (C3H or WT). A fold change <1 indicates an increase and <1 indicates decrease. Means and p-values are detailed in (Table S2). Color indicates direction of change – increase in WD (red), decrease in WD (blue), and lipids not detected (black). Color intensity indicates p-values – dark (p < 0.05 and FDR-adjusted p < 0.1) and light (p ≥ 0.05 or FDR-adjusted p ≥ 0.1). (*) denotes lipids correlating with oxidative stress marker 9-HETE (r ≥ 0.4 and <0.6, p ≤ 0.05, FDR-adjusted p ≤ 0.1). C3H n = 12 and tx-j n = 18; WT n = 25 and Atp7b−/− B6 n = 23. Atp7b−/− B6, The Atp7b global knockout on a C57Bl/6 background; Auto, non-enzymatic auto-oxidation; C3H, C3HeB/FeJ control mice; COX, cyclooxygenase; CYP, cytochrome P450 monooxygenase; DH, ehydrogenase; DiHDoPA, dihydroxydocosapentaenoic acid; DiHDPE, dihydroxydocosapentaenoic acid; DiHETE, dihydroxyeicosatetraenoic acid; DiHETrE, dihydroxyeicosatrienoic acid; DiHO, dihydroxyoctadecanoic acid; DiHODE, dihydroxyoctadecadienoic acid; DiHOME, dihydroxyoctadecenoic acid; Elov, fatty acid elongase; Ep-KODE, epoxyoxooctadecenoic acid; EpETE, epoxyeicosatetraenoic acid; EpETrE, EpO, Epoxystearic acid; Epoxyeicosatrienoic acid; EpODE, epoxyoctadecadienoic acid; EpOME, epoxyoctadecenoic acid; Fads, fatty acid desaturase; HDoHE, hydroxydocosahexaenoic acid; HEPE, hydroxyeicosapentaenoic acid; HETE, hydroxyeicosatetraenoic acid; HODE, hydroxyoctadecadienoic acid; HOTE, hydroxyoctadecatrienoic acid; HpODE, hydroperoxyoctadecadienoic acid; KETE, keto-eicosatetraenoic; KODE, keto-octadecadienoic acid; LOX, lipoxygenase; PGE, prostaglandin E; PGF, prostaglandin F; sEH, soluble epoxide hydrolase; TriHOME, trihydroxyoctadecaenoic acid; TXB2, thromboxane B2; tx-j, The toxic milk mice from The Jackson Laboratory; WT, Atp7b+/+ controls.
