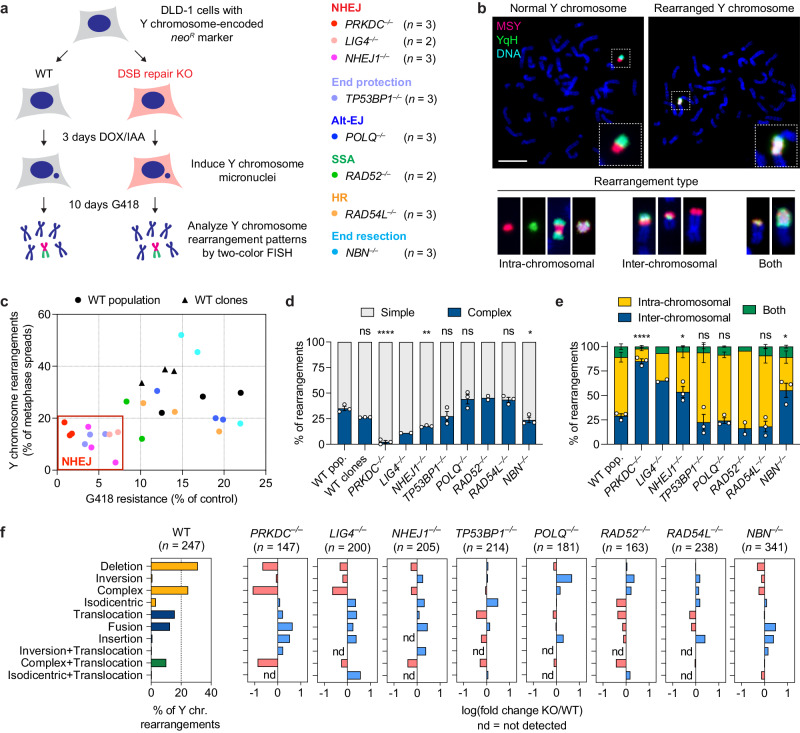
Fig. 1. Genomic rearrangement landscape of mis-segregated chromosomes in the absence of specific DNA double-strand break (DSB) repair pathways.
a Experimental approach to survey the impact of specific DSB repair pathways on chromosome rearrangements induced by micronucleus formation. Biallelic gene knockouts (KOs) were generated in the background of the CEN-SELECT system in isogenic DLD-1 cells. Y chromosome-specific mis-segregation into micronuclei and rearrangements were induced by treatment with doxycycline and auxin (DOX/IAA). b Representative examples of metaphase spreads with normal or derivative Y chromosomes. Different types of rearrangements can be visualized by DNA fluorescence in situ hybridization (FISH) using probes targeting the euchromatic portion of the male-specific region (MSY, red) and the heterochromatic region (YqH, green) of the Y chromosome. Rearrangements were induced by 3d DOX/IAA treatment followed by G418 selection. Scale bar, 10 µm. c Plot summarizing the effect on cell viability after G418 selection (x-axis) and rearrangement frequency of the Y chromosome (y-axis) for each DSB repair KO clone. d Proportion of Y chromosomes exhibiting simple or complex rearrangements, as determined by metaphase FISH, following transient centromere inactivation. e Proportion of inter- and/or intra-chromosomal rearrangements. Data in (d) and (e) represent the mean ± SEM of n = 3 independent experiments for WT population, n = 2 KO clones for LIG4 and RAD52, and n = 3 KO clones for WT, DNA-PKcs, XLF, 53BP1, POLQ, RAD54, and NBS1; statistical analyses were calculated by ordinary one-way ANOVA test with multiple comparisons. In (d), ns not significant; ****p < 0.0001; **p = 0.0015; *p = 0.0456. In (e), ns not significant; ****p < 0.0001; *p = 0.0229 (NHEJ1−/−) or 0.0142 (NBN−/−). f Left: distribution of Y chromosome rearrangement types as determined by metaphase FISH following 3d DOX/IAA treatment and G418 selection. Data are pooled from three independent experiments. Right: plots depict the mean fold change in each rearrangement type as compared to WT cells. Sample sizes indicate the number of rearranged Y chromosomes examined; data are pooled from two or three individual KO clones per gene. See also Supplementary Fig. 2. Source data are provided as a Source Data file.