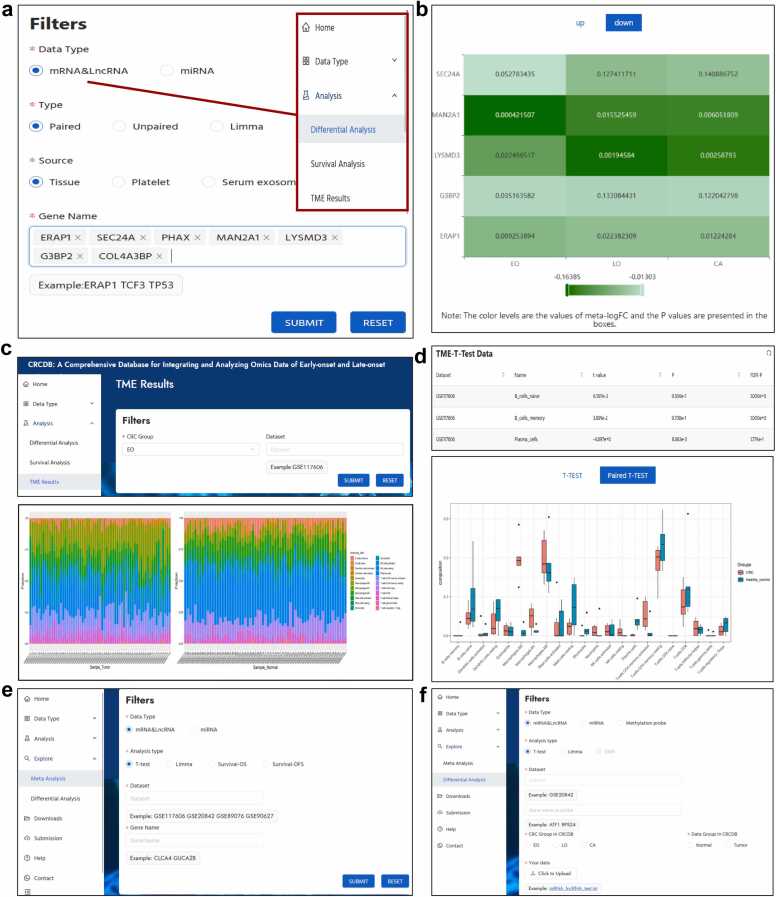
Fig. 3.
Analysis module of CRCDB. a,b. The meta-analysis results of differential analysis using RRA. The color levels of the heatmap are in accordance with the meta-FC values and the P values are presented in the boxes. c. The 22 immune cells fraction in each sample of the selected dataset. d. The differential analysis results of the immune cell fraction between tumor and normal samples in both table and bar chart form. e. In the Explore-Meta analysis part, users could choose the datasets of their interest and get the new meta-analysis results in the heatmap format. f. On the Explore-differential analysis page, users could upload the transcriptome data or the methylation data and do the differential analysis in combination with the data in the CRCDB.