Highlights
-
•
ETV4 expression is increased in CCA.
-
•
ETV4 facilitates the proliferation, migration, and invasion of CCA cells through regulating the TGF-β signaling.
-
•
ETV4 enhances the glycolysis of CCA cells through regulating the TGF-β signaling.
Keywords: TGF-β/Smad2/3 signaling pathway, ETV4, Cholangiocarcinoma
Abstract
Background
Considerable studies show that ETS variant 4 (ETV4) plays an important roles in multitudinous tumor. This study investigated its function in cholangiocarcinoma (CCA) progression and revealed the underlying mechanisms.
Methods
The expression of ETV4 in CCA was evaluated using TCGA database and the single-cell analysis based on GSE189903 dataset. ETV4 expression in CCA human specimens was detected by reverse transcription-quantitative PCR, immunohistochemistry, and western blot. Cell Counting Kit-8, EdU, colony formation, wound healing, and Transwell assays were used to analyze the effects of ETV4. Extracellular acidification rate, oxygen consumption rate, glucose uptake, and lactate production were used to measure glycolysis in CAA cells. Western blot was performed to explore glycolysis-related proteins. Tumor growth was evaluated in mice xenograft tumors.
Results
ETV4 was up-regulated in CCA epithelial cells. The high-expression of ETV4 was associated with poor prognosis of patients with CCA. ETV4 overexpression enhanced the proliferation, migration, invasion, and glycolysis of CCA cells; ETV4 silencing led to the contrary effects. Mechanistically, ETV4 activates TGF-β/Smad2/3 signaling pathway. In mice xenograft mode, ETV4 silencing inhibits the tumor growth, the expression of glycolysis-related proteins and TGF-β/Smad2/3 pathway proteins.
Conclusions
ETV4 functions as an essential factor in the roles of TGF-β1 in CCA cells, and may be a promising target for TGF-β1-mediated CCA progression.
Introduction
Cholangiocarcinoma (CCA) is an uncommon cancer in the bile ducts [1]. CCA is linked to high mortality rate [2]. Since no obvious symptoms in early stage CCA, no distinct risk factors, and lacking standardized program for cancer diagnosis, patients receives late treatment and has a poor prognosis [3]. Surgical resection offers the only potential chance of recovery from CCA [4]. However, surgical resection could be performed in 20 %−30 % cases of CCA [4]. Numerous genes with altered expression have been identified in patients with CCA compared to healthy people [5]. In the last few years, the advancements in targeted therapies have increased the treatment option to patients with CCA [6]. Therefore, having a deeper comprehending of the biological process of CCA will help researchers in exploring targeted treatment strategies for patients suffering from CCA. Further discovery of therapeutic targets are needed to enhance patient survival rate.
ETS variant 4 (ETV4), a member of the ETS family polyomavirus enhancer activator 3 subfamily, that plays critical roles in multiple malignancies [[7], [8], [9]]. ETV4 aggravates metastasis of pancreatic ductal adenocarcinoma via activating the CXCL13/CXCR5 signaling [10]. ETV4 regulates the activity of estrogen receptor alpha and the progression of endometrial cancer [11]. ETV4 potentiates the progression of hepatocellular carcinoma through activation of YAP [12], TNF-α [13], and ANXA2 [14]. ETV4 controls the malignant progression of gastric cancer through mediating KDM5D [15]. To date, the roles of ETV4 in CCA still unclear.
Transforming growth factor β (TGF-β) signaling participates in plentiful biological processes, including proliferation, differentiation, and migration [16]. In early-stage cancer cells, TGF-β signaling exists tumor-suppressor functions by inducing cell-cycle arrest and stimulating apoptosis. On the contrary, in late-stage cancer cells, TGF-β promotes metastasis of cancer cells [17]. The cascade system among TGF-β signaling and other signaling pathways, including glycolysis, enriches our knowledge of its functions in cancer [[18], [19], [20]]. In glioblastoma cells, TGF-β1 promotes glycolysis via inducing PFKFB3 by stimulating p38 MAPK and PI3K/Akt signaling [21] TGF-β induces glycolysis and promotes migration in pancreatic cancer cells [22]. In lung cancer cells, ANGPTL2 potentiates glycolysis through activating TGF-β-ZEB1-GLUT3 axis [23]. Owing to extensive effects of TGF-β, restraint of TGF-β gives numerous therapeutic prospects. Hence, the future expectations for therapy that inhibits the TGF-β signaling in CCA appears bright.
In the study, we describe the effects of ETV4 on CCA cells. We declined ETV4 expression and induced overexpression of ETV4 in CCA cells, and determined to study whether ETV4 regulated CCA cells through the TGF-β signaling. The findings of this study will provide a therapeutic target in CCA.
Materials and methods
Single-cell analysis
The GSE189903 single-cell dataset was download from Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/). A total of 14 tissue samples were filtered for data analysis, including 11 CCA tumor tissue samples and 3 paracancer tissue samples. The single-cell analysis was carried out using the R package Seurat (version 5.0.3) [24] in the R program (version 4.3.0) [25]. Firstly, cells with UMI counts less than 200 or more than 5000, mitochondrial content greater than 5 % and features greater than 1500 were filtered out. Use the ‘NormalizeData’ function for data normalization, scale. factor=10,000. The ‘FindVariableGenes’ function and vst method were used to identify variable genes and select the first 2000 features. The ‘ScaleData’ function was used to standardize and centralize the features. Secondly, ‘RunPCA’ function was used for principal component analysis, and 50 principal components were retained. ‘JackStraw’ function was used to determine the significance of PCA score. Using R package harmony (version 1.2.0) [26] to integrate all sample data. Cell clusters were identified using the ‘FindClusters’ function (resolution=0.8) based on the shared nearest neighbor (SNN) modular optimization clustering algorithm. ‘RunUMAP’ function was used to reduce the Uniform Manifold Approximation and Projection (UMAP) dimension and the ‘DimPlot’ function was used for visualization. Then, using the CellMarker2.0 database [27] and artificially collated reported marker genes to annotate cell clusters. Finally, ETV4 gene expression matrix annotated as bile duct epithelial cells was extracted. The significance difference of ETV4 gene expression between Tumour group and Normal group was analyzed using R packet rstatix (version 0.7.2) by the T test. Hypothesis test correction method was Benjamini-Hochberg. The analysis results were visualized as bar graphs using the R package ggplot2 (version 3.5.0) [28].
TCGA database analysis
TCGA database is utilized to analyze ETV4 expression in CCA. Overall survival was predicted using GEPIA with ETV4 expression in CHOL-TCGA. The differential genes identified based on CHOL-TCGA data were enriched and analyzed by HALLMARK gene set and KEGG pathway. Gene Set Enrichment Analysis (GSEA) was conducted using HALLMARK and KEGG gene sets.
Human specimens
Human CCA and adjacent tissues (n = 27) were collected from patients who underwent tumor resection between December 2020 and November 2022 at our hospital. The CCA and normal tissues were stored at −80 °C. This study was collected with informed consent in writing and approved by the Institutional Ethics Committee of our hospital.
Histological analysis
For histological analysis, tumor were fixed, embedded, and sectioned into slices (5-μm). The sections were deparaffinized by xylene at room temperature for 10 min, rehydrated by ethanol gradient for 2 min, and blocked endogenous peroxidase activity by 3 % H2O2 at room temperature for 10 min. After blocking with blocking solution, sections were incubated with anti-ETV4 (1:200, 10,684–1-AP, Proteintech) and anti-Ki67 (1:200, ab15580, Abcam) overnight at 4 °C. After washing, sections were incubated with secondary antibody and the color was developed using 3,3′ diaminobenzidine tetrahydrochloride and hematoxylin. Images were measured using a microscope (Nikon, Japan). The histological score was calculated by multiplying the intensity and positivity scores. The staining intensity was scored as follows: 0–negative, 1–weak, 2–medium or 3–strong. The staining positivity, i.e., the percentage of positive staining areas of tumour cells in relation to the whole tumour area, was scored as follows: 0 (0 %), 1 (1–25 %), 2 (26–50 %), 3 (51–75 %) and 4 (76–100 %). The staining was evaluated by two pathologists who blinded to this study.
Cell culture
Human CCA cell lines RBE (CL-0191, Procell, Wuhan, China), HuCCT1 (CL-0725, Procell), and HCCC9810 (CL-0095, Procell) were maintained in RPMI-1640 medium with 10 % fetal bovine serum (FBS; HyClone, USA). CCLP1 cell lines (MZ-2377, MINGZHOUBIO, Ningbo, China) were cultured in DMEM medium with 10 % FBS (HyClone). Cells were cultured at 37 °C with 5 % CO2.
Cell transfection
siRNA was utilized to knock down the ETV4 in HuCCT1 and HCCC9810 cells. pcDNA3.1-ETV4 was used to elevate ETV4 expression in RBE cells. The siRNAs and pcDNA3.1-ETV4 were synthesized by RiboBio (Guangzhou, China). The sequence of negative control siRNA (si-NC) was as follows: 5′–UUCUCCGAACGUGUCACGUTT–3′, 5′–ACGUGACACGUUCGGAGAATT–3′. The sequence of si1-ETV4 was: 5′–CCGAUACUAUUAUGAGAAAGG–3′, 5′ –UUUCUCAUAAUAGUAUCGGAG–3′. The sequence of si2-ETV4 was: 5′–GCAGAGCUUUAAGCAAGAAUA–3′, 5′–UUCUUGCUUAAAGCUCUGCUG–3′. Transfections of the siRNAs and pcDNA3.1-ETV4 were performed by using Lipofectamine 2000 (ThermoFisher Scientific, USA). For in vivo experiments, HuCCT1 cells were transfected with sh-ETV4 by the lentivirus (RiboBio, China) and screened with puromycin for 14 days. The sequence of sh-ETV4 was as follows: 5′–GGTGGTGATCAAACAGGAA–3′, 5′–TTCCTGTTTGATCACCACC–3′. The sequence of sh-NC was as follows: 5′–TTCTCCGAACGTGTCACGTTT-3′, 5′–ACGTGACACGTTCGGAGAATT–3′.
Cell counting kit‑8 (CCK-8)
The cells in 96-well plate were incubated at 37 °C for 1, 2, 3, and 4 days. At each time-point, CCK-8 solution (10 μl; C0037, Beyotime) was added to each well and incubated at 37 °C for 2 h. Microplate reader (Bio-Rad, USA) was utilized to read the absorbance at 450 nm.
EdU
Cells were incubated in 24-well plates (2 × 104) for 24 h. EdU reagent (Beyotime) was added to cells for 2 h. Cells were fixed with 4 % paraformaldehyde and stained by DAPI. Images were acquired using a fluorescence microscope.
Colony formation
Cells were incubated in 6-well plates (800 cells/well) for 2 weeks. Cells were fixed with methanol for 15 min and stained with 0.1 % crystal violet (Sigma-Aldrich) for 20 min. Colonies was counted with a microscope.
Would healing assay
CCA cells grown to monolayer in 6-well plates. Monolayer was wounded using a 200 μl pipette tip. Cells were cultured for 24 h. Images were captured at 0 h and 24 h using a microscope and wound size was measured using Image J software.
Transwell assay
A sample of 2 × 105 cells in serum-free medium were plated into the upper chamber of Transwell system (Corning, USA). Culture media with 10 % FBS was added to the lower chambers. After incubation for 24 h, cells on the lower surface were fixed with methanol, stained with 0.1 % crystal violet for 20 min, and counted with a microscope. For the invasion, the upper chamber was pre-coated with Matrigel (BD Biosciences).
Cellular respiration and glycolytic activity
Extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) were detected using Seahorse XF Cell Mito Stress Test kit and Seahorse XF Glycolysis Stress Test kit (Agilent Technologies, USA) on the Seahorse XF96 analyzer (Seahorse). Briefly, transfected cells (1 × 104) were seeded into a Seahorse XF 96 microplate for 10 h and measured ECAR and OCR. For ECAR, glucose, oligomycin, and 2-DG were sequentially added into each well at 30, 60, 90 min. For OCR, oligomycin, FCCP, and Rote/AA were sequentially added [29].
Glucose uptake and lactate production
The glucose uptake levels was detected using a Glucose Uptake Assay Kit (ab136955, Abcam). The lactate production levels was detected using a lactate assay kit (MAK064, Solarbio, China).
Quantitative reverse transcription PCR (qRT-PCR)
Total RNA was extracted from the CCA tissues and cells using Beyozol (R0011, Beyotime). The cDNA was synthesized using HiScript II Q Select RT SuperMix (R223–01, Vazyme, China). qRT-PCR was performed using ChamQ SYBR qPCR Master Mix (Q311–02, Vazyme) with the Applied Biosystems system 7500 (Thermo Fisher Scientific). The following primers for genes: ETV4 5′–GAAAAACAAGTCGGTGCGCT–3′, 5′–TTTCCGGGCGATTTCTGAGG–3′. β-actin, 5′–AACACCCCAGCCATGTACGTT–3′, 5′–CCATCTCTTGCTCGAAGTCCA–3′.
Western blot
CCA tissues and cells were lysed with RIPA (Beyotime). Proteins were separated by SDS-PAGE gels and transferred to membranes (Millipore). After blocking, membranes were incubated with anti-ETV4 (1:1000, 10,684–1-AP, Proteintech), hexokinase-II (HK2; 1:1000, ab209847, Abcam), pyruvate kinase M2 (PKM2; 1:1000, 15,822–1-AP, Proteintech), lactate dehydrogenase A (LDHA; 1:2000, 19,987–1-AP, Proteintech), Vimentin (1:2000, 10,366–1-AP, Proteintech), N-cadherin (1:2000, 22,018–1-AP, Proteintech), E-cadherin (1:20,000, 20,874–1-AP, Proteintech), and β-actin (1:5000, 81,115–1-RR, Proteintech) at 4 °C overnight, and then incubated with anti-Rabbit IgG (1:10,000, SA00001–2, Proteintech) for 1 h at room temperature. The membranes were detected by enhanced chemiluminescence.
Xenograft mice models
Female BALB/c nude mice (6–8 weeks) were purchased from GemPharmatech Co., Ltd. (Nanjing, China). Mice were randomly assigned into two groups (n = 5). After a week of adaptive feeding, 5 × 106 stable transfected HuCCT1 cells were subcutaneously injected into the flanks of mice. Tumor diameters were measured every three days. Volume (mm3) = (length × width 2)/2. After 31 days, tumors were excised, imaged, and weighed.
TUNEL staining
The tumor tissue sections were fixed in 4 % paraformaldehyde for 30 min, washed with PBS for twice, and permeabilized in 0.3 % Triton X-100 at room temperature for 5 min. After incubation with 0.3 % H2O2 for 20 min, the tissue sections were stained using a TUNEL Apoptosis Assay Kit (Beyotime, Shanghai, China). The TUNEL-stained cells were captured under a fluorescence microscope (Nikon, Japan) and counted.
Statistical analysis
Data are shown as mean ± standard deviation. Prism version 7.0 software was used for statistical analysis. Comparisons between two groups were carried out using unpaired t-test. One-way ANOVA followed by Tukey's post-hoc test or two-way ANOVA followed by Bonferroni's post-hoc test were used for multiple‑group comparisons. P < 0.05 was considered statistically significant.
Results
Increased expression of ETV4 in CCA
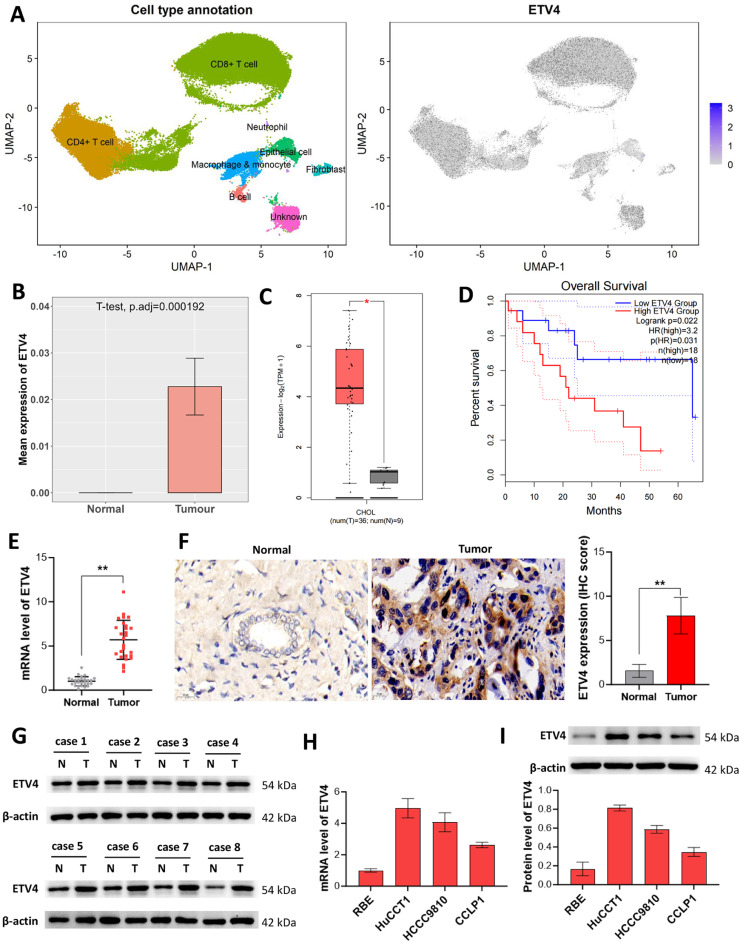
The result of single-cell analysis using GSE189903 dataset showed that, ETV4 was extensively expressed in CD8+ T cells, CD4+ T cells, macrophages & monocytes, fibroblasts, B cells, and epithelial cells (Fig. 1A). Compared to normal group, ETV4 was highly expressed in tumor epithelial cells (Fig. 1B). Also, TCGA data showed that ETV4 was upregulated in tumor tissues (Fig. 1C) and its high-expression is associated with the predicted poor survival of patients with CCA (Fig. 1D). Next, we examined ETV4 expression in the collected human tissues. ETV4 mRNA expression was higher in CCA tissues than the paired normal tissues (Fig. 1E). Consistent results were showed from IHC and western blot results (Fig. 1F and G). Furthermore, we analyzed ETV4 expression in four CCA cell lines through qRT-PCR and western blot analysis (Fig. 1H and I). HuCCT1 and HCCC9810 cells with higher expression of ETV4 and RBE cells with lower expression of ETV4 were selected for use in the following.
Fig. 1.
Increased expression of ETV4 in CCA (A) Single-cell analysis of ETV4 expression in CCA using GSE189903 dataset. (B) ETV4 expression in tumor epithelial cells of single-cell analysis results. (C) TCGA data analysis of ETV4 expression in CCA and normal tissues. (D) Kaplan‑Meier analysis of survival curve of patients with ETV4 low expression and ETV4 high expression based on TCGA data. (E-G) ETV4 expression in CCA tissues was detected using qRT-PCR (E), IHC (F), and western blot (G). (H and I) ETV4 level in CCA cell lines was detected by qRT-PCR (H) and western blot (I). Data were analyzed using the unpaired t-test. *p < 0.05 and **p < 0.01 compared with Normal group.
ETV4 depletion decreases the growth of CCA cells
To elucidate the role of ETV4 in CCA, cells were transfected with si1-ETV4, si2-ETV4, pcDNA3.1-ETV4, or the negative controls. si1-ETV4 and si2-ETV4 transfection markedly knocked down ETV4 expression in HuCCT1 and HCCC9810 cells. pcDNA3.1-ETV4 transfection significantly increased ETV4 expression in RBE cells (Fig. 2A). ETV4 knockdown significantly declined cell viability, In contrast, ETV4 overexpression elevated cell viability (Fig. 2B). ETV4 silencing decreased the rate of EdU-positive cells, whereas ETV4 overexpression significantly enhanced the rate of EdU-positive cells (Fig. 2C). In addition, a decreased colony number upon ETV4 knockdown was observed in HuCCT1 and HCCC9810 cells, whereas an increased colony number upon ETV4 overexpression was observed in RBE cells (Fig. 2D). Above findings indicated that ETV4 knockdown suppressed the proliferation of CCA cells.
Fig. 2.
ETV4 depletion decreases the growth of CCA cells. (A) Western blot analysis of ETV4 expression after transfection. (B-D) CCK-8 assay (B), EdU (C), and clone formation (D) for detecting cell proliferation after transfection. Data in (B) were analyzed using the two-way ANOVA analysis followed by Bonferroni's post-hoc test. Data in (C and D) were analyzed using the one-way ANOVA analysis followed by Tukey's post-hoc test. **p < 0.01 compared with si-NC or vector group.
ETV4 depletion decreases the migration and invasion of CCA cells
ETV4 knockdown significantly suppressed scratch healing, whereas ETV4 overexpression promoted scratch healing (Fig. 3A). ETV4 knockdown significantly reduced migration and invasion of CCA cells; whereas, ETV4 overexpression markedly induced migration and invasion of CCA cells (Fig. 3B). In addition, ETV4 knockdown decreased Vimentin and N-cadherin expression, and increased E-cadherin expression; whereas, ETV4 overexpression lead to the opposite regulation on these proteins (Fig. 3C). Thus, the migration and invasion in CCA cells was mediated by ETV4.
Fig. 3.
ETV4 depletion decreases the migration and invasion of CCA cells. (A) Would healing for CAA cell migration after transfection. (B) Transwell assay for CAA cell migration and invasion after transfection. (C) Western blot for detecting the expression of Vimentin, N-cadherin, and E-cadherin in CAA cells. Data were analyzed using the one-way ANOVA analysis followed by Tukey's post-hoc test. **p < 0.01 compared with si-NC or vector group.
ETV4 depletion decreases the glycolysis of CCA cells
Increased glycolysis potentiates cell proliferation by meeting energy requirements of cancer cells [30]. We evaluated ECAR, a mark of overall glycolytic flux and OCR, which reflects mitochondrial oxidative respiration. We found that ETV4 knockdown declined ECAR and elevated OCR in HuCCT1 and HCCC9810 cells, whereas ETV4 overexpression elevated ECAR and declined OCR in RBE cells (Fig. 4A and B). Silencing of ETV4 decreased glucose uptake and lactate production levels in HuCCT1 and HCCC9810 cells. In contrast, ETV4 overexpression increased glucose uptake and lactate production levels in RBE cells (Fig. 4C and D). To further investigate the roles of ETV4 in glycolysis, we detected key enzymes in glycolysis. The expression of HK2, PKM2, and LDHA was declined in HuCCT1 and HCCC9810 cells with ETV4 knockdown, while was enhanced in RBE cells with ETV4 overexpression (Fig. 4E). Overall, above results indicated that silencing of ETV4 inhibited the glycolysis of CCA cells.
Fig. 4.
ETV4 depletion decreases the glycolysis of CCA cells. (A and B) Extracellular acidification rate (ECAR; A) and oxygen consumption rate (OCR; B) were measured using the Seahorse XF96 analyzer. (C and D) Glucose uptake (C) and lactate production (D) were used to measure glycolysis in CAA cells. (E) Western blot analysis of the expression of HK2, PKM2, and LDHA in CAA cells. Data were analyzed using the one-way ANOVA analysis followed by Tukey's post-hoc test. *p < 0.05 and **p < 0.01 compared with si-NC or vector group.
ETV4 depletion inhibits the TGF-β signaling in CCA cells
The molecular mechanism was further elucidated by HALLMARK gene set and KEGG pathway enrichment analysis. HALLMARK gene set enrichment analysis revealed the enrichment of TGF-β and glycolysis (Fig. 5A). GSEA with hallmark gene sets showed that ETV4 expression was linked to glycolysis in CHOL-TCGA dataset (Fig. 5B). KEGG pathway enrichment analysis instructed an enrichment of TGF-β (Fig. 5C). GSEA with KEGG gene sets found an enrichment of TGF-β signaling in CHOL-TCGA tissues with high ETV4 expression (Fig. 5D). Thus, we appraised the effects of ETV4 on TGF-β activation. ETV4 knockdown decreased TGF-β1 expression and Smad2/3 phosphorylation, while ETV4 overexpression increased TGF-β1 expression and Smad2/3 phosphorylation (Fig. 5E). Thus, we inferred that upregulation of ETV4 is essential for TGF-β1/Smad2/3 to promote glycolysis-induced tumor cell growth (Fig. 5F).
Fig. 5.
ETV4 depletion inhibits the TGF-β signaling in CCA cells. (A) The bubble chart of enriched pathway using HALLMARK enrichment analysis. (B) GSEA with hallmark gene sets showed that ETV4 expression was linked to glycolysis in CHOL-TCGA dataset. (C) The bubble chart of enriched pathway using KEGG enrichment analysis. (D) GSEA with KEGG gene sets showed enrichment of TGF-β signaling in CHOL-TCGA tissues with high ETV4 expression (E) Western blot analysis of the expression of TGF-β1 and Smad2/3 in CAA cells. (F) Schematic diagram of ETV4-mediated glycolysis promotion in CAA cell growth.
ETV4 depletion decreases the growth, migration, and invasion of CCA cells via regulating the TGF-β signaling
To test our hypothesis, we added TGF-β1 and its inhibitor SB525334 into the culture medium of HuCCT1 and RBE cells, respectively. EdU and colony formation results showed that TGF-β1 significantly eliminated the inhibitory effect of ETV4 knockdown on the proliferation of HuCCT1 cells. In RBE cells, SB525334 partly reversed ETV4 overexpression-mediated promotion of proliferation (Fig. 6A and B). Treatment with TGF-β1 reversed ETV4 knockdown-reduced migration and invasion. Treatment with SB525334 reversed ETV4 overexpression-induced migration and invasion (Fig. 6C). Overall, these findings specified that ETV4 enhanced proliferation, migration, and invasion of CCA cells by regulating the TGF-β signaling.
Fig. 6.
ETV4 depletion decreases the growth, migration, and invasion of CCA cells via regulating the TGF-β signaling. HuCCT1 cells were treated with TGF-β1 and RBE cells were treated with TGF-β1 inhibitor SB525334. (A and B) EdU (A) and colony formation (B) assays for detecting cell proliferation after the indicated treatment. (C) Transwell assay for cell migration and invasion after treatment. Data were analyzed using the one-way ANOVA analysis followed by Tukey's post-hoc test. *p < 0.05 and **p < 0.01 compared to the indicated group.
ETV4 depletion decreases the glycolysis of CCA cells via regulating the TGF-β signaling
Next, we investigate whether ETV4 regulates glycolysis in CCA cells by mediating the TGF-β signaling. Treatment with TGF-β1 reversed the effects of ETV4 knockdown on ECAR, OCR, glucose uptake, and lactate production. Treatment with SB525334 reversed the effects of ETV4 overexpression on ECAR, OCR, glucose uptake, and lactate production (Fig. 7A-D). Furthermore, TGF-β1 partly neutralized ETV4 knockdown-reduced HK2, PKM2, and LDHA expression, whereas SB525334 reversed ETV4 overexpression-induced HK2, PKM2, and LDHA expression (Fig. 7E). Together, these data indicated that ETV4 mediated glycolysis of CCA cells via the TGF-β signaling.
Fig. 7.
ETV4 depletion decreases the glycolysis of CCA cells via regulating the TGF-β signaling. HuCCT1 cells were treated with TGF-β1 ad RBE cells were treated with TGF-β1. (A and B) ECAR (A) OCR (B), glucose uptake (C), and lactate production (D) were measured to evaluate glycolysis in CAA cells. (E) Western blot analysis of the expression of HK2, PKM2, and LDHA in CAA cells. Data were analyzed using the one-way ANOVA analysis followed by Tukey's post-hoc test. *P < 0.05 and **p < 0.01 compared to the indicated group.
ETV4 depletion inhibits the oncogenicity of CCA cells in vivo
To further confirm above results, we subcutaneously injected HuCCT1 cells into nude mice. The expression of ETV4 in the excised tumours was detected for confirming the shRNA in vivo efficacy. Results in Fig. 8A showed that, as compared to the sh-NC group, ETV4 was lower expressed in sh-ETV4 group. ETV4 knockdown reduced the growth of xenograft tumors (Fig. 8B-D). ETV4 knockdown in vivo markedly inhibited the cholangiocyte overgrowth, as the Ki67-positive cell rate was reduced and the TUNEL-positive cell rate was increased (Fig. 8E). Moreover, the expression of HK2, PKM2, and LDHA was markedly decreased in xenograft tumors by ETV4 knockdown (Fig. 8F). TGF-β1 expression and Smad2/3 phosphorylation were declined in xenograft tumors upon ETV4 knockdown (Fig. 8G). Thus, above results elucidated that ETV4 knockdown suppressed the oncogenicity of CCA cells in vivo.
Fig. 8.
ETV4 depletion inhibits the oncogenicity of CCA cells in vivo. (A) Western blot analysis of the expression of ETV4 in the excised tumor tissues. (B) Tumor images at day 31. (C) Cell growth curve of tumor with control or ETV4 knockdown. (D) Tumor weight at day 31. (E) Ki67 and TUNEL staining of tumor sections. (F) Western blot analysis of the expression of HK2, PKM2, and LDHA in tumor tissues with control or ETV4 knockdown. (G) Western blot analysis of the expression TGF-β1 and p-Smad2/3 in tumor tissues with control or ETV4 knockdown. Data in (C) were analyzed using two-way ANOVA followed by Bonferroni's post-hoc test. Others were analyzed using the unpaired t-test. **p < 0.01 compared with sh-NC group.
Discussion
In this research, we elaborated that ETV4 markedly promoted the growth, migration, invasion, and glycolysis by activating the TGF-β/Smad2/3 signaling, which confirmed the carcinogenic effects of ETV4 on CCA.
ETV4 is highly expressed in plentiful cancers and its high expression is linked to poor outcomes [[31], [32], [33]]. Yao et al. elucidated that ETV4 is highly expressed in colon adenocarcinoma and indicated advanced stage in patients [32]. Dumortier et al. revealed that ETV4 is associated with worse survival of patients with breast cancer [33]. Cheng et al. observed the up-regulation of ETV4 in lung adenocarcinoma [34]. Our research data from TCGA and tissues collected from patients with CCA showed that ETV4 expression was elevated in CCA tissues as compared to the adjacent normal tissues. In vitro, ETV4 obviously increased proliferation, migration and invasion of CCA cells. In vivo, the volume of tumor was decreased upon ETV4 knockdown. These results was agreed with previous studies that ETV4 facilitates the progression of breast [35], lung [36], pancreatic [37], and papillary thyroid cancers [38].
Cancer cells take advantage of glycolysis for proliferation [39]. HK2 is an enzyme that catalyzes the phosphorylation of hexose [40]. It is the first enzyme in the glycolysis and also the rate-limiting enzyme in the glycolysis [41]. PKM2 is dominating exist in proliferating cells with abundant anabolic requirements, such as embryo and tumors [42]. LDHA catalyzes the conversion of pyruvate to lactate [43]. These enzymes are overexpressed in tumors and are responsible for supporting tumor growth [44]. Restraining the function of these enzymes may blockade glucose utilization and could be an excellent therapeutic option for inhibiting cancer progression [45]. Jing et al. reported that NCAPD3 interacts with c-Myc to increase glycolytic regulators, and stimulates glycolysis, and finally boosts progression of colorectal cancer [46]. Wu et al. instructed that SETD1A and HIF1α together accelerated glycolysis and aggravate progression of gastric carcinoma [47]. To assess the role of ETV4 in glucose utilization in CCA cells, we analyzed ECAR, OCR, glucose uptake, and lactate production, and glycolytic enzymes in ETV4 knockdown and overexpression CCA cells. Silencing of ETV4 reduced ECAR, elevated OCR, diminished glucose uptake and lactate production levels, and declined HK2, PKM2, and LDHA expression in HuCCT1 and HCCC9810 cells. These opposite results were observed in RBE cells with ETV4 overexpression. Similarly, ETV4 facilitates breast cancer cell stemness through stimulating glycolysis and sonic hedgehog pathway [35]. Overall, above results indicated that silencing of ETV4 inhibited glycolysis.
Important evidence has confirmed that TGF-β/SMAD2/3 signaling pathway is critical for progression of cancer [[48], [49], [50]]. circPTEN1 suppresses colorectal cancer progression by blockading TGF-β/Smad2/3 signaling [48]. LINC00941 promotes colorectal cancer metastasis by restraining degradation of SMAD4 protein and stimulating the TGF-β/SMAD2/3 signaling [51]. In pancreatic cancer, itraconazole suppresses invasion and migration of tumor cells through refraining TGF-β/SMAD2/3 signaling [52]. In gastric cancer, ginsenoside Rh4 inhibits metastasis of cells via SIX1-TGF-β/Smad2/3 signaling [53]. We reported that TGF-β/Smad2/3 signaling was activated with ETV4 overexpression. Treatment with TGF-β1 abrogated the inhibitory effects of ETV4 knockdown on progression of CCA. Treatment with SB525334 reversed ETV4 overexpression-induced progression of CCA. Consistently, ETV4 facilitates progression of liver cancer through mediating the TGF-β signaling, whereas SB525334 treatment profoundly refrained the roles of ETV4 [54]. TGF-β induces cell metabolism into glycolysis and facilitates tumor growth [55]. Knockdown of Smad2/3 and HIF-1α block TGF-β-induced tumor growth without oxygen [56]. TGF-β modulates glycolysis primarily by PKM2 [56]. Cheng et al. found that metformin blocks TGF-β1-mediated migration through reducing mTOR signaling to decline PKM2 expression in cervical carcinoma cells [57]. Xu et al. shown that PKCε potentiates growth of prostate cancer cells through enhancing aerobic glycolysis by cascading Smad2/3 [58]. In this study, TGF-β1 partly annulled ETV4 knockdown-reduced glycolysis, whereas SB525334 reversed ETV4 overexpression-induced glycolysis.
In summary, ETV4 promoted the progression of CCA by regulating glycolysis via the TGF-β signaling. Our study suggested that ETV4 may be a therapeutic potential for CCA.
Funding
Shandong Province medical health science and technology project. (202304010282).
Availability of data and material
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The experimental protocol of our study was performed in accordance with the Guide for the Care and Use of Laboratory Animals and approved by Shandong Provincial Hospital Affiliated to Shandong First Medical University. All study participants gave their informed consent. The experiments were approved by the Research Ethics Board of Shandong Provincial Hospital Affiliated to Shandong First Medical University.
CRediT authorship contribution statement
Fangfeng Liu: Writing – review & editing, Writing – original draft, Conceptualization. Qianchang Wang: Software, Resources. Zhengjian Wang: Visualization, Supervision, Resources. Shizhe Zhang: Project administration, Methodology. Qingqiang Ni: Validation, Investigation, Formal analysis. Hong Chang: Resources, Investigation, Formal analysis.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Not applicable
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2024.102035.
Appendix. Supplementary materials
References
- 1.Brown Z.J., Ruff S.M., Pawlik T.M. Developments in FGFR and IDH inhibitors for cholangiocarcinoma therapy. Expert. Rev. AntiCancer Ther. 2023;23(3):257–264. doi: 10.1080/14737140.2023.2176846. [DOI] [PubMed] [Google Scholar]
- 2.Testa U., Pelosi E., Castelli G. Cholangiocarcinoma: molecular abnormalities and cells of origin. Technol. Cancer Res. Treat. 2023;22(15330338221128689) doi: 10.1177/15330338221128689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Izquierdo-Sanchez L., Lamarca A., La Casta A., Buettner S., Utpatel K., Klümpen H.J., Adeva J., Vogel A., Lleo A., Fabris L., et al. Cholangiocarcinoma landscape in Europe: diagnostic, prognostic and therapeutic insights from the ENSCCA Registry. J. Hepatol. 2022;76(5):1109–1121. doi: 10.1016/j.jhep.2021.12.010. [DOI] [PubMed] [Google Scholar]
- 4.Moris D., Palta M., Kim C., Allen P.J., Morse M.A., Lidsky M.E. Advances in the treatment of intrahepatic cholangiocarcinoma: an overview of the current and future therapeutic landscape for clinicians. CA Cancer J. Clin. 2023;73(2):198–222. doi: 10.3322/caac.21759. [DOI] [PubMed] [Google Scholar]
- 5.Wang S., Yu L., Sun X., Zhang B. Establishment and verification of potential biomarkers for cholangiocarcinoma. Exp. Ther. Med. 2022;24(3) doi: 10.3892/etm.2022.11483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Elvevi A., Laffusa A., Scaravaglio M., Rossi R.E., Longarini R., Stagno A.M., Cristoferi L., Ciaccio A., Cortinovis D.L., Invernizzi P., et al. Clinical treatment of cholangiocarcinoma: an updated comprehensive review. Ann. Hepatol. 2022;27(5):7. doi: 10.1016/j.aohep.2022.100737. [DOI] [PubMed] [Google Scholar]
- 7.Xie M., Lin Z., Ji X., Luo X., Zhang Z., Sun M., Chen X., Zhang B., Liang H., Liu D., et al. FGF19/FGFR4-mediated elevation of ETV4 facilitates hepatocellular carcinoma metastasis by upregulating PD-L1 and CCL2. J. Hepatol. 2023;79(1):109–125. doi: 10.1016/j.jhep.2023.02.036. [DOI] [PubMed] [Google Scholar]
- 8.Zheng C., Liu M., Ge Y., Qian Y., Fan H. HBx increases chromatin accessibility and ETV4 expression to regulate dishevelled-2 and promote HCC progression. Cell Death. Dis. 2022;13(2):022–04563. doi: 10.1038/s41419-022-04563-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Q., Liu S., Wang H., Xiao K., Lu J., Chen S., Huang M., Xie R., Lin T., Chen X. ETV4 mediated tumor-associated neutrophil infiltration facilitates Lymphangiogenesis and lymphatic metastasis of bladder cancer. Adv. Sci. 2023;10(11):20. doi: 10.1002/advs.202205613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao X., Jiang M., Chu Y., Han Y., Jin Y., Zhang W., Wang W., Yang S., Li W., Fan A., et al. ETV4 promotes pancreatic ductal adenocarcinoma metastasis through activation of the CXCL13/CXCR5 signaling axis. Cancer Lett. 2022;524:42–56. doi: 10.1016/j.canlet.2021.09.026. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez A.C., Vahrenkamp J.M., Berrett K.C., Clark K.A., Guillen K.P., Scherer S.D., Yang C.H., Welm B.E., Janát-Amsbury M.M., Graves B.J., et al. ETV4 Is necessary for estrogen signaling and growth in endometrial cancer cells. Cancer Res. 2020;80(6):1234–1245. doi: 10.1158/0008-5472.CAN-19-1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu X., Wang B., Liu Y., Jing T., Xu G., Zhang L., Kun J., Chen Z., Xiang L., Xu C., et al. ETV4 potentiates nuclear YAP retention and activities to enhance the progression of hepatocellular carcinoma. Cancer Lett. 2022;537(215640):13. doi: 10.1016/j.canlet.2022.215640. [DOI] [PubMed] [Google Scholar]
- 13.Qi D., Lu M., Xu P., Yao X., Chen Y., Gan L., Li Y., Cui Y., Tong X., Liu S., et al. Transcription factor ETV4 promotes the development of hepatocellular carcinoma by driving hepatic TNF-α signaling. Cancer Commun. 2023;5(10):12482. doi: 10.1002/cac2.12482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun T., Zhang J. ETV4 mediates the Wnt/β-catenin pathway through transcriptional activation of ANXA2 to promote hepatitis B virus-associated liver hepatocellular carcinoma progression. J. Biochem. 2021;170(5):663–673. doi: 10.1093/jb/mvab088. [DOI] [PubMed] [Google Scholar]
- 15.Cai L.S., Chen Q.X., Fang S.Y., Lian M.Q., Lian M.J., Cai M.Z. ETV4 promotes the progression of gastric cancer through regulating KDM5D. Eur. Rev. Med. Pharmacol. Sci. 2020;24(5):2442–2451. doi: 10.26355/eurrev_202003_20511. [DOI] [PubMed] [Google Scholar]
- 16.Zhao M., Mishra L., Deng C.X. The role of TGF-β/SMAD4 signaling in cancer. Int. J. Biol. Sci. 2018;14(2):111–123. doi: 10.7150/ijbs.23230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Colak S., Ten Dijke P. Targeting TGF-β Signaling in Cancer. Trends. Cancer. 2017;3(1):56–71. doi: 10.1016/j.trecan.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 18.Jena B.C., Das C.K., Banerjee I., Bharadwaj D., Majumder R., Das S., Biswas A., Kundu M., Roy P.K., Kundu C.N., et al. TGF-β1 induced autophagy in cancer associated fibroblasts during hypoxia contributes EMT and glycolysis via MCT4 upregulation. Exp. Cell Res. 2022;417(1):11. doi: 10.1016/j.yexcr.2022.113195. [DOI] [PubMed] [Google Scholar]
- 19.Torrealba N., Vera R., Fraile B., Martínez-Onsurbe P., Paniagua R., Royuela M. TGF-β/PI3K/AKT/mTOR/NF-kB pathway. Clinicopathological features in prostate cancer. Aging Male. 2020;23(5):801–811. doi: 10.1080/13685538.2019.1597840. [DOI] [PubMed] [Google Scholar]
- 20.Zhuang X., Zhang H., Li X., Cong M., Peng F., Yu J., Zhang X., Yang Q., Hu G. Differential effects on lung and bone metastasis of breast cancer by Wnt signalling inhibitor DKK1. Nat. Cell Biol. 2017;19(10):1274–1285. doi: 10.1038/ncb3613. [DOI] [PubMed] [Google Scholar]
- 21.Rodríguez-García A., Samsó P., Fontova P., Simon-Molas H., Manzano A., Castaño E., Rosa J.L., Martinez-Outshoorn U., Ventura F., Navarro-Sabaté À., et al. TGF-β1 targets Smad, p38 MAPK, and PI3K/Akt signaling pathways to induce PFKFB3 gene expression and glycolysis in glioblastoma cells. FEBS. J. 2017;284(20):3437–3454. doi: 10.1111/febs.14201. [DOI] [PubMed] [Google Scholar]
- 22.Costanza B., Rademaker G., Tiamiou A., De Tullio P., Leenders J., Blomme A., Bellier J., Bianchi E., Turtoi A., Delvenne P., et al. Transforming growth factor beta-induced, an extracellular matrix interacting protein, enhances glycolysis and promotes pancreatic cancer cell migration. Int. J. Cancer. 2019;145(6):1570–1584. doi: 10.1002/ijc.32247. [DOI] [PubMed] [Google Scholar]
- 23.Osumi H., Horiguchi H., Kadomatsu T., Tashiro K., Morinaga J., Takahashi T., Ikeda K., Ito T., Suzuki M., Endo M., et al. Tumor cell-derived angiopoietin-like protein 2 establishes a preference for glycolytic metabolism in lung cancer cells. Cancer Sci. 2020;111(4):1241–1253. doi: 10.1111/cas.14337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hao Y., Stuart T., Kowalski M.H., Choudhary S., Hoffman P., Hartman A., Srivastava A., Molla G., Madad S., Fernandez-Granda C. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat. Biotechnol. 2024;42(2):293–304. doi: 10.1038/s41587-023-01767-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Team RC R: a language and environment for statistical computing. MSOR Connect. 2014;1 [Google Scholar]
- 26.Korsunsky I., Millard N., Fan J., Slowikowski K., Raychaudhuri S. harmony: fast, sensitive, and accurate integration of single cell data. R package version. 2021;1 doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu C., Li T., Xu Y., Zhang X., Li F., Bai J., Chen J., Jiang W., Yang K., Ou Q., et al. CellMarker 2.0: an updated database of manually curated cell markers in human/mouse and web tools based on scRNA-seq data. Nucleic Acids Res. 2022;51(D1):D870–D876. doi: 10.1093/nar/gkac947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villanueva R.A.M., Chen Z.J. Taylor & Francis; 2019. ggplot2: Elegant Graphics For Data Analysis. [Google Scholar]
- 29.Li L., Liang Y., Kang L., Liu Y., Gao S., Chen S., Li Y., You W., Dong Q., Hong T., et al. Transcriptional regulation of the warburg effect in cancer by SIX1. Cancer Cell. 2018;33(3):368–385. doi: 10.1016/j.ccell.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 30.Vander Heiden M.G., Cantley L.C., Thompson C.B. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324(5930):1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang R., Peng Y., Gao Z., Qian J., Yang K., Wang X., Lu W., Zhu Y., Qiu D., Jin T., et al. Oncogenic role and drug sensitivity of ETV4 in human tumors: a pan-cancer analysis. Front. Oncol. 2023;13(1121258) doi: 10.3389/fonc.2023.1121258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao D., Bao Z., Qian X., Yang Y., Mao Z. ETV4 transcriptionally activates HES1 and promotes Stat3 phosphorylation to promote malignant behaviors of colon adenocarcinoma. Cell Biol. Int. 2021;45(10):2129–2139. doi: 10.1002/cbin.11669. [DOI] [PubMed] [Google Scholar]
- 33.Dumortier M., Ladam F., Damour I., Vacher S., Bièche I., Marchand N., de Launoit Y., Tulasne D. Chotteau-Lelièvre A: ETV4 transcription factor and MMP13 metalloprotease are interplaying actors of breast tumorigenesis. Breast. Cancer Res. 2018;20(1):018–0992. doi: 10.1186/s13058-018-0992-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cheng T., Zhang Z., Cheng Y., Zhang J., Tang J., Tan Z., Liang Z., Chen T., Liu Z., Li J., et al. ETV4 promotes proliferation and invasion of lung adenocarcinoma by transcriptionally upregulating MSI2. Biochem. Biophys. Res. Commun. 2019;516(1):278–284. doi: 10.1016/j.bbrc.2019.06.115. [DOI] [PubMed] [Google Scholar]
- 35.Zhu T., Zheng J., Zhuo W., Pan P., Li M., Zhang W., Zhou H., Gao Y., Li X., Liu Z. ETV4 promotes breast cancer cell stemness by activating glycolysis and CXCR4-mediated sonic hedgehog signaling. Cell Death. Discov. 2021;7(1):021–00508. doi: 10.1038/s41420-021-00508-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y., Ding X., Liu B., Li M., Chang Y., Shen H., Xie S.M., Xing L., Li Y. ETV4 overexpression promotes progression of non-small cell lung cancer by upregulating PXN and MMP1 transcriptionally. Mol. Carcinog. 2020;59(1):73–86. doi: 10.1002/mc.23130. [DOI] [PubMed] [Google Scholar]
- 37.Tyagi N., Deshmukh S.K., Srivastava S.K., Azim S., Ahmad A., Al-Ghadhban A., Singh A.P., Carter J.E., Wang B., Singh S. ETV4 facilitates cell-cycle progression in pancreatic cells through transcriptional regulation of cyclin D1. Mol. Cancer Res. 2018;16(2):187–196. doi: 10.1158/1541-7786.MCR-17-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L., Zhang Y., Yang J., Liu L., Yao B., Tian Z., He J. The knockdown of ETV4 inhibits the papillary thyroid cancer development by promoting ferroptosis upon SLC7A11 downregulation. DNA Cell Biol. 2021;40(9):1211–1221. doi: 10.1089/dna.2021.0216. [DOI] [PubMed] [Google Scholar]
- 39.Lunt S.Y., Vander Heiden M.G. Aerobic glycolysis: meeting the metabolic requirements of cell proliferation. Annu Rev. Cell Dev. Biol. 2011;27:441–464. doi: 10.1146/annurev-cellbio-092910-154237. [DOI] [PubMed] [Google Scholar]
- 40.Li L., Zhang X., Lin Y., Ren X., Xie T., Lin J., Wu S., Ye Q. Let-7b-5p inhibits breast cancer cell growth and metastasis via repression of hexokinase 2-mediated aerobic glycolysis. Cell Death. Discov. 2023;9(1):023–01412. doi: 10.1038/s41420-023-01412-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan V.P., Miyamoto S. HK2/hexokinase-II integrates glycolysis and autophagy to confer cellular protection. Autophagy. 2015;11(6):963–964. doi: 10.1080/15548627.2015.1042195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z., Deng X., Liu Y., Sun L., Chen F. PKM2, function and expression and regulation. Cell Biosci. 2019;9(52):019–0317. doi: 10.1186/s13578-019-0317-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miao P., Sheng S., Sun X., Liu J., Huang G. Lactate dehydrogenase A in cancer: a promising target for diagnosis and therapy. IUBMB Life. 2013;65(11):904–910. doi: 10.1002/iub.1216. [DOI] [PubMed] [Google Scholar]
- 44.Long W., Gong X., Yang Y., Yang K. Downregulation of PER2 promotes tumor progression by enhancing glycolysis via the phosphatidylinositol 3-Kinase/Protein kinase B pathway in oral squamous cell carcinoma. J. Oral Maxillofac. Surg. 2020;78(10):1. doi: 10.1016/j.joms.2020.05.035. [DOI] [PubMed] [Google Scholar]
- 45.Deng C., Li H., Li Q. F-box protein 17 promotes glioma progression by regulating glycolysis pathway. Biosci. Biotechnol. Biochem. 2022;86(4):455–463. doi: 10.1093/bbb/zbac008. [DOI] [PubMed] [Google Scholar]
- 46.Jing Z., Liu Q., He X., Jia Z., Xu Z., Yang B., Liu P. NCAPD3 enhances Warburg effect through c-myc and E2F1 and promotes the occurrence and progression of colorectal cancer. J. Exp. Clin. Cancer Res. 2022;41(1):022–02412. doi: 10.1186/s13046-022-02412-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu J., Chai H., Xu X., Yu J., Gu Y. Histone methyltransferase SETD1A interacts with HIF1α to enhance glycolysis and promote cancer progression in gastric cancer. Mol. Oncol. 2020;14(6):1397–1409. doi: 10.1002/1878-0261.12689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng L., Liang H., Zhang Q., Shen Z., Sun Y., Zhao X., Gong J., Hou Z., Jiang K., Wang Q., et al. circPTEN1, a circular RNA generated from PTEN, suppresses cancer progression through inhibition of TGF-β/Smad signaling. Mol. Cancer. 2022;21(1):022–01495. doi: 10.1186/s12943-022-01495-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bertrand-Chapel A., Caligaris C., Fenouil T., Savary C., Aires S., Martel S., Huchedé P., Chassot C., Chauvet V., Cardot-Ruffino V., et al. SMAD2/3 mediate oncogenic effects of TGF-β in the absence of SMAD4. Commun. Biol. 2022;5(1):022–03994. doi: 10.1038/s42003-022-03994-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bertero A., Brown S., Madrigal P., Osnato A., Ortmann D., Yiangou L., Kadiwala J., Hubner N.C., de Los Mozos I.R., Sadée C., et al. The SMAD2/3 interactome reveals that TGFβ controls m(6)A mRNA methylation in pluripotency. NatureNature. 2018;555(7695):256–259. doi: 10.1038/nature25784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu N., Jiang M., Liu H., Chu Y., Wang D., Cao J., Wang Z., Xie X., Han Y., Xu B. LINC00941 promotes CRC metastasis through preventing SMAD4 protein degradation and activating the TGF-β/SMAD2/3 signaling pathway. Cell Death. Differ. 2021;28(1):219–232. doi: 10.1038/s41418-020-0596-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen K., Cheng L., Qian W., Jiang Z., Sun L., Zhao Y., Zhou Y., Zhao L., Wang P., Duan W., et al. Itraconazole inhibits invasion and migration of pancreatic cancer cells by suppressing TGF-β/SMAD2/3 signaling. Oncol. Rep. 2018;39(4):1573–1582. doi: 10.3892/or.2018.6281. [DOI] [PubMed] [Google Scholar]
- 53.Jiang H., Ma P., Duan Z., Liu Y., Shen S., Mi Y., Fan D. Ginsenoside Rh4 Suppresses Metastasis of Gastric Cancer via SIX1-Dependent TGF-β/Smad2/3 Signaling Pathway. Nutrients. 2022;14(8) doi: 10.3390/nu14081564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou Z., Wu B., Chen J., Shen Y., Wang J., Chen X., Fei F., Li L. ETV4 facilitates proliferation, migration, and invasion of liver cancer by mediating TGF-β signal transduction through activation of B3GNT3. Genes. Genomics. 2023;31(10):023–01428. doi: 10.1007/s13258-023-01428-z. [DOI] [PubMed] [Google Scholar]
- 55.Gong L., Ji L., Xu D., Wang J., Zou J. TGF-β links glycolysis and immunosuppression in glioblastoma. Histol. Histopathol. 2021;36(11):1111–1124. doi: 10.14670/HH-18-366. [DOI] [PubMed] [Google Scholar]
- 56.Huang Y., Chen Z., Lu T., Bi G., Li M., Liang J., Hu Z., Zheng Y., Yin J., Xi J., et al. HIF-1α switches the functionality of TGF-β signaling via changing the partners of smads to drive glucose metabolic reprogramming in non-small cell lung cancer. J. Exp. Clin. Cancer Res. 2021;40(1):021–02188. doi: 10.1186/s13046-021-02188-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cheng K., Hao M. Metformin Inhibits TGF-β1-induced epithelial-to-mesenchymal transition via PKM2 Relative-mTOR/p70s6k signaling pathway in cervical carcinoma cells. Int. J. Mol. Sci. 2016;17(12) doi: 10.3390/ijms17122000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu W., Zeng F., Li S., Li G., Lai X., Wang Q.J., Deng F. Crosstalk of protein kinase C ε with Smad2/3 promotes tumor cell proliferation in prostate cancer cells by enhancing aerobic glycolysis. Cell Mol. Life Sci. 2018;75(24):4583–4598. doi: 10.1007/s00018-018-2914-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.