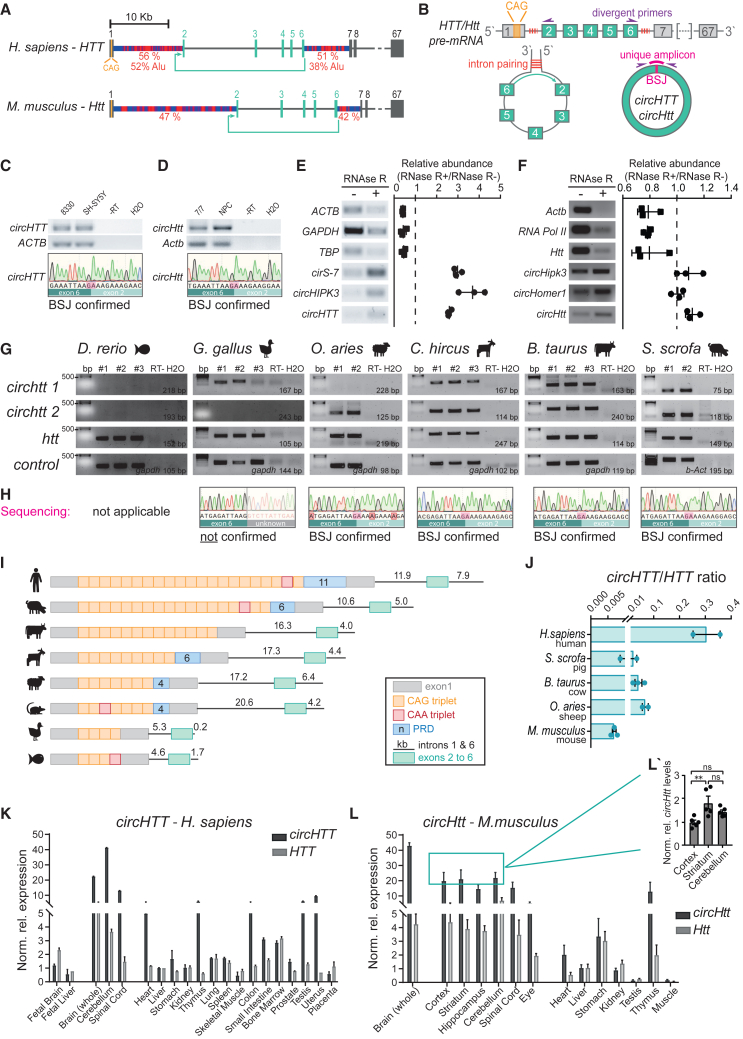
Figure 1.
Identification and validation of the evolutionary conserved circHTT(2,3,4,5,6)
(A) Schematic representation of the Huntington’s disease gene locus in humans and mice. Circularized exons in turquoise; CircHTT(2,3,4,5,6)/circHtt(2,3,4,5,6): 484 nt, exons 2–6; % of nucleotides accounting for repetitive sequences in flanking introns in red. (B) Schematic of putative circHTT(2,3,4,5,6)/circHtt(2,3,4,5,6) biogenesis and amplification of the back-splice junction (BSJ) using divergent primers. (C and D) Representative images of gel electrophoresis after PCR amplification (top) and electropherograms after sequencing (bottom) of the human (hiPSC-derived neural progenitors 8330-8, and neuroblastoma SH-SY5Y cells) and mouse (neuro-progenitor and striatal StHdh Q7/7 cells) amplicons, spanning the BSJ. (E and F) RNase R (+) and buffer only (−) treated total RNA was purified and used for cDNA synthesis, followed by endpoint PCR for linear and circRNA targets and gel electrophoresis; left: representative gel images; right: quantification of relative abundance (n = 3 biological replicates in E and F, respectively, data are plotted as mean ± standard error of the mean (SEM). (G) Gel electrophoresis of amplicons from endpoint PCR on cDNA synthetized from total RNA of brain samples of different vertebrate species. Two divergent primer pairs were designed targeting the predicted circHTT(2,3,4,5,6) orthologue sequences (top) of each selected species, as well as primers against linear HTT and housekeeping gene orthologues (GAPDH or ACTB) (n = 2/3 biological replicates per species). (H) Gel extraction and Sanger sequencing of the longer amplicons from predicted circHTT(2,3,4,5,6) orthologues in (G). (I) Schematic representation of the CAG tract of HTT exon 1 as well as the length of introns 1 and 6 (flanking exons 2 and 6) of the selected species. (J) RT-qPCR analysis of circHTT(2,3,4,5,6) orthologue expression levels reported as a ratio from the linear HTT orthologue RNA levels in brain samples (circHTT/HTT ratio was calculated upon normalization to GAPDH housekeeping gene). Species are organized according to the length of their CAG tract (decreasing order, n = 2/3 brain samples per species, data are plotted as mean ± SEM). (K and L) Total RNA from the human tissue panel (K, Ambion, #AM6000) and adult wild-type (C57BL6J) mouse tissues/body district (L, n = 3 mice) was analyzed by qPCR for circHTT(2,3,4,5,6) and HTT, and circHtt(2,3,4,5,6), Htt levels respectively (human transcripts were normalized on NONO, mouse transcripts on Pgk1 levels and shown relative to transcript expression levels in liver, data are plotted as mean ± SEM). (L′) Direct comparison between circHtt(2,3,4,5,6) expression levels in the adult cortex, cerebellum, and striatum (n = 5 mice, one-way ANOVA with multiple comparisons testing, ∗∗p < 0.01, data are plotted as mean ± SEM).