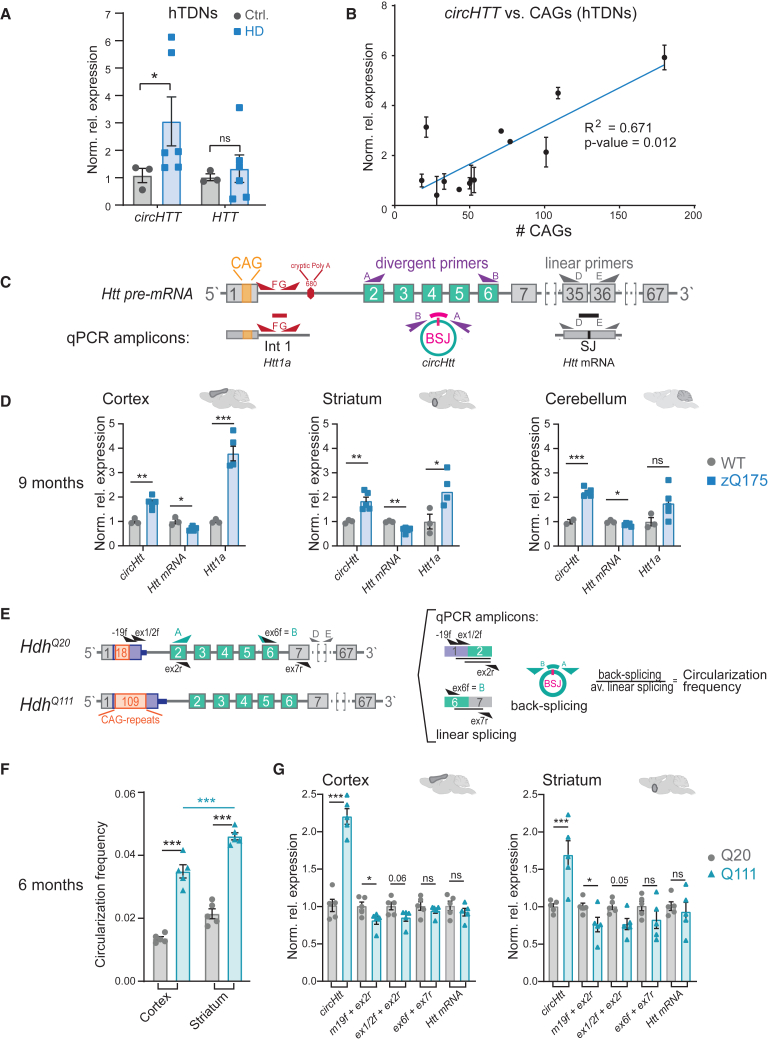
Figure 2.
Correlation between circHTT (2,3,4,5,6)/circHtt(2,3,4,5,6) levels and CAG trinucleotide repeat number
(A and B) CircHTT(2,3,4,5,6) expression levels assessed by RT-qPCR on induced pluripotent stem cell (iPSC)-derived terminal differentiated cortical neurons (hTDNs) from controls (n = 3) and HD patients (n = 6/8, Mann-Whitney test in (A), ∗∗p < 0.01. (B) Results of a linear regression; data are plotted as mean ± SEM). (C) Scheme of qRT-PCR strategy to assess expression levels of different transcripts (i.e., toxic fragment Htt1a, circHtt(2,3,4,5,6) and linear Htt mRNA) from the Htt locus in the brains of wild-type and the zQ175 knockin mouse model for HD. (D) RT-qPCR on brain samples from indicated brain regions (cortex, striatum, cerebellum) of 9-month-old wild-type and zQ175 mice (n = 3 wild-type and 5 zQ175 biological replicates per tissue, one-way ANOVA with Sidak's multiple comparisons testing, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05; data are plotted as mean ± SEM). (E) Experimental strategy to test circHtt(2,3,4,5,6) back-splicing frequency-schematic representation of the binding sites of the employed primer pairs (top). CircHtt fw primer, indicated with letter B in the scheme, can also be used to detect linear Htt when combined with ex7 rv primer. (F) Circularization frequency in cortical, striatal, and cerebellar samples of 6-month-old Q20 (n = 5) and Q111 (n = 5) mice (the relative level of expression of circHtt(2,3,4,5,6) and linear isoforms was first calculated normalizing on the Pgk1 housekeeping gene and subsequently the circularization frequency—as ratio between back-splicing and linear splicing—was computed; one-way ANOVA with Sidak's multiple comparisons testing, ∗∗∗p < 0.001; data are plotted as mean ± SEM). (G) Normalized relative expression of individual primer sets used to calculate circularization frequencies in 6-month Q20 and Q111 mouse tissues (2−DDCt values, unpaired two-sided t tests for each primer set individually, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, ns = not significant).