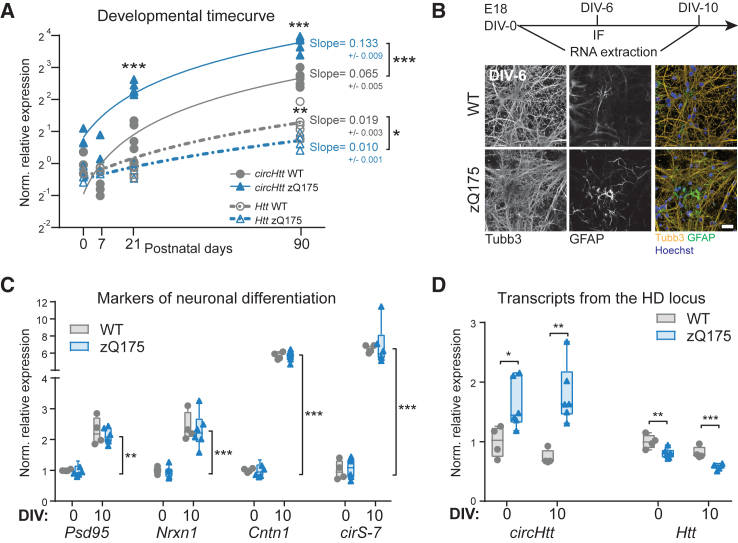
Figure 3.
In vivo and in vitro developmental trajectory of circHtt(2,3,4,5,6) abundance in the zQ175 HD mouse model
(A) RT-qPCR analysis across postnatal (P) development in the wild-type and zQ175 mouse striatum (A, n(P0) = 3 per genotype, n(P7) = 3 wild-type/2 zQ175, n(P21) = 5 per genotype, n(P90) = 5 per genotype; transcript levels normalized to the geometric mean of three housekeeping genes, Pgk1, Tfrc, and Actb, and to the wild-type levels at P0; two-way ANOVAs followed by Tukey's multiple comparisons test, and simple linear regression to calculate expression trajectory slopes, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05). (B) Primary neuronal culture experimental layout from wild-type and zQ175 E18 embryos (top). Representative images of primary cortical neurons at 6 days in vitro (DIV), stained for Tubb3+ differentiating neurons and GFAP+ astrocytes (bottom), scale bar, 25 μm; (C and D) RT-qPCR results on cDNA from total RNA of cultured neurons at 0 and 10 DIV; neuronal maturation markers Psd95, Nrxn1, Cntn1, and the circular RNA cirS-7 (C); circHtt(2,3,4,5,6) and Htt mRNA levels in the zQ175 derived neurons at 0 DIV and 10 DIV (D) (n(embryos) = 4 wild-type and 5 zQ175; transcript levels normalized to the geometric mean of three housekeeping genes, Pgk1, Tfrc, and Actb, and to the wild-type levels at DIV-0; two-way ANOVA followed by Tukey's multiple comparisons test, ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05; data are plotted as mean ± SEM).