Figure 4.
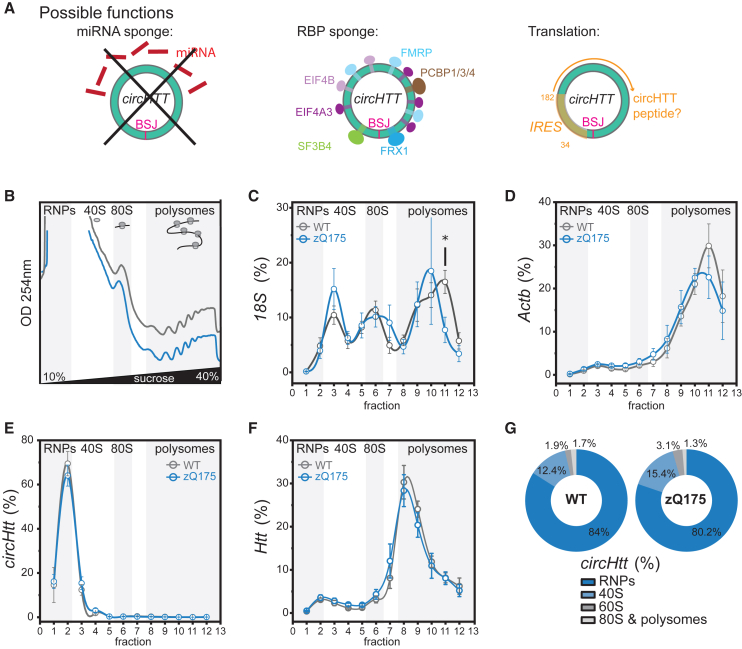
CircHTT(2,3,4,5,6) has predicted binding sites for RNA-binding proteins (RBPs) and associates with the 40S ribosomal subunit
(A) Sequence analysis of circHTT(2,3,4,5,6) reveals no enrichment in miRNA binding sites (left); however, binding sites for multiple RBPs (middle, Table S2, width visualizes number of nucleotides in binding motive), as well as an IRES sequence (right, localized between nucleotides 31 and 128), followed by a predicted open reading frame of 186 aa. (B) Representative sucrose gradient absorbance profiles of polysome fractionation of cytoplasmic lysates from wild-type (WT) (gray) and zQ175 (blue) brain samples at 6–7 months of age. (C–F) Relative distribution of 18S, Actb, circHtt(2,3,4,5,6), and Htt RNAs along polysome profile in WT (gray line) and zQ175 (blue line) (data are plotted as mean ± SEM of n = 5 independent biological replicates). (G) Doughnut plots reporting the relative percentages of circHtt(2,3,4,5,6) co-sedimentation with RNPs, 40S, 60S, 80S, and polysomes.