Figure 5.
Overexpression of circHtt(2,3,4,5,6) ameliorates cellular phenotypes of the STHdh Q111/Q111 striatal cell model system for HD
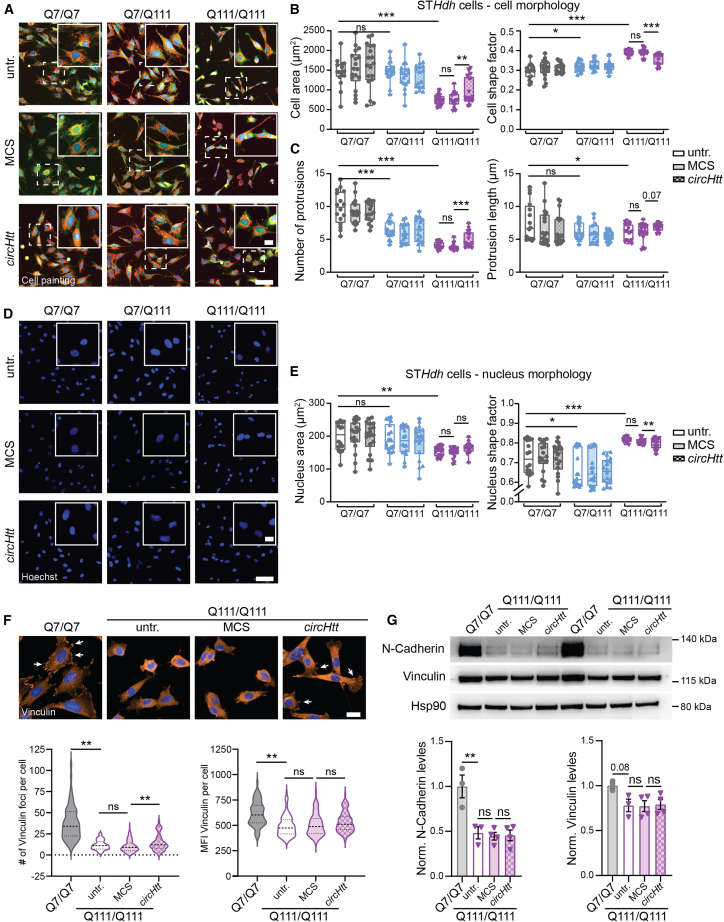
(A) Representative images of the high-throughput cell painting assay using Calcein-AM (green), MitoTracker (red), and Hoechst (blue) live-cell staining to assess cellular features of the untransfected (untr.) control STHdh Q7/Q7, Q7/Q111, and Q111/Q111 cell lines, as well as MCS and circHtt(2,3,4,5,6) overexpressing cells of all genotypes (composite merged image of the three channels is shown in A, close ups of the dash-boxed cells in the upper right corner). (B) Average cell area in μm2 (left) and cell shape factor (0 = flat, 1 = perfect circle, right) are reported. (C) Average number (left) and length (right) of cellular protrusions. (D) Representative images of Hoechst nuclear staining. (E) Average nuclear cell area in μm2 (left), and shape factor (0 = flat, 1 = perfect circle, right); (parameters as calculated upon segmentation of Calcein-AM and Hoechst signal by the Custom Module extension of MetaXpress (6.7.2.290) are reported; (A–E, n(cells) = 8,000–10,000 cells per genotype and condition over three biological replicates, each dot represents the average value of all cells from the individual replica wells; outliers were removed using the ROUT (Q = 1%) method, followed by one-way ANOVA with Sidak's multiple comparisons testing (parametric data) and Kruskal-Wallis with Dunn's multiple comparisons testing (nonparametric data), ∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05, scale bars indicate 100 μm in overview and 25 μm in close ups; data are plotted as mean ± SEM). (F) Immunofluorescence staining of Q7/Q7 wild-type control and Q111/Q111 untransfected, MCS, and circHtt(2,3,4,5,6) overexpressing cells for Vinculin (orange) and Hoechst (blue) (top). The average number of Vinculin foci per cell (bottom left) and average Vinculin fluorescence intensity (bottom right) as quantified by an automated cell segmentation and analysis Macro in Fiji (n(technical replicates) = 3, with n(cells) = 69 Q7/Q7, 102 Q111/Q111 untransfected, 134 Q111/111 MCS, and 95 Q111/Q111 circHtt(2,3,4,5,6) overexpressing; outliers were removed using the ROUT (Q = 1%) method, followed by Kruskal-Wallis with Dunn's multiple comparisons testing [nonparametric data], ∗∗p < 0.01, ns = not significant, scale bars indicate 20 μm; data are presented with violin plots, central line indicates median, dotted line quartiles). (G) Representative western blots of N-Cadherin and Vinculin levels in Q7/Q7 wild-type control and Q111/Q111 untransfected, MCS, and circHtt(2,3,4,5,6) overexpressing cells (top) and quantifications normalized to Hsp90 (bottom) (n(biological replicates) = 3–4 per genotype and condition, one-way ANOVA with Sidak's multiple comparisons testing, ∗∗p < 0.01, ns = not significant; data are plotted as mean ± SEM).