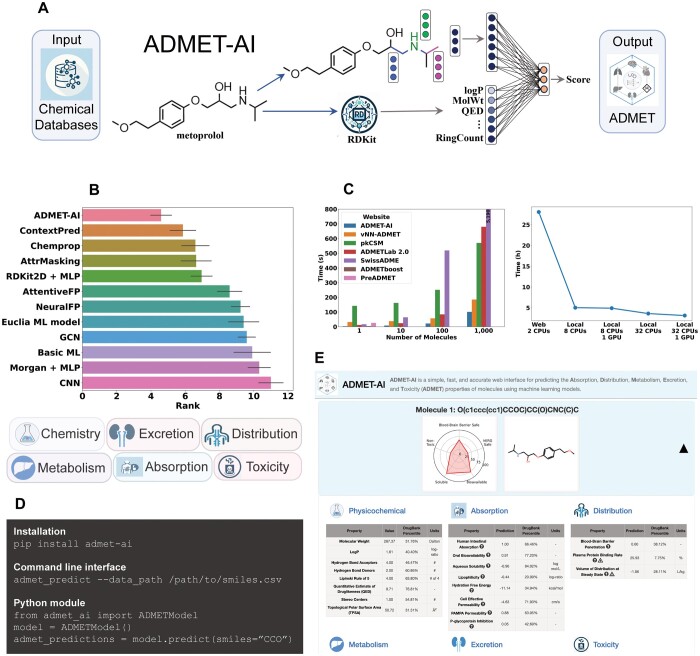
Figure 1.
Overview of ADMET-AI. (A) An illustration of training an ADMET-AI graph neural network Chemprop-RDKit model. (B) The overall rank of ADMET-AI models on the Therapeutics Data Commons ADMET leaderboard of 22 ADMET datasets. Representative overall categories predicted by ADMET-AI are shown below. Error bars indicate standard error across datasets. (C) The computational efficiency of ADMET-AI. Left panel, the time (in seconds, median of three trials) for the ADMET-AI web server and other common ADMET web servers to make predictions on 1, 10, 100, or 1000 molecules from the DrugBank reference set. ADMETboost and PreADMET are limited to one molecule. Since pkCSM and SwissADME are limited to 100 and 200 molecules, respectively, their 1000 molecule times are computed as 100 molecule time . Right panel, the time (in hours, median of three trials) for the ADMET-AI web server and various hardware configurations running the ADMET-AI local command line tool to make predictions on 1 million molecules from the DrugBank reference set (1000 copies of the 1000 molecule DrugBank set). Since the ADMET-AI web server is currently limited to 1000 molecules, its 1 million molecule time is computed as 1000 molecule time . (D) Commands needed to install and run the local version of ADMET-AI, either as a command line tool or as a Python module. (E) Predictions displayed on the ADMET-AI website (admet.ai.greenstonebio.com).