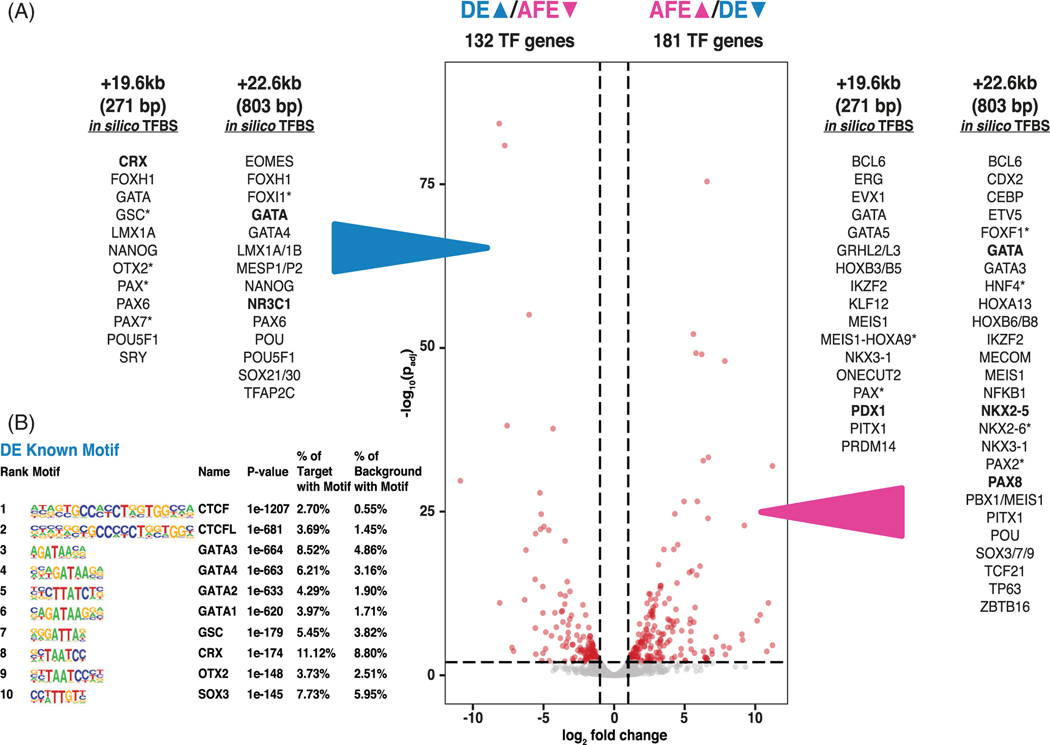
FIGURE 2.
In silico predicted transcription factor binding sites (TFBS) at +19.6 kb and +22.6 kb cis-regulatory elements (CREs) correspond to definitive endoderm (DE) > anterior foregut endoderm (AFE) differentially expressed TFs. A, Volcano plot of weighted fold change as a function of P-values for TF genes during DE to AFE transition. DEGs passing a fold change of 2 and adjusted P value of .01 are shown in red. In silico TFBS predictions for the +19.6 kb and +22.6 kb CREs were performed using MatInspector and transcription factor affinity prediction (TRAP). Predicted binding sites for DE differentially upregulated TFs (left) and AFE differentially upregulated TFs (right) are listed (data are from MatInspector unless marked *, which denotes TRAP; bold font denotes predictions from both analyses). B, HOMER analysis showing top 10 overrepresented binding motifs for known transcription factors under open chromatin peaks in DE cells (adapted from Reference 25)