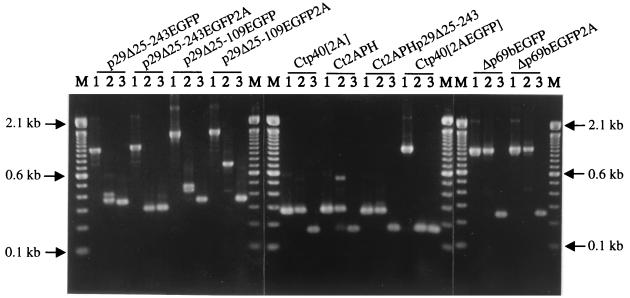
FIG. 4.
ClampR analysis of dsRNA recovered from fungal colonies transfected with candidate expression vectors. For each replication-competent vector (shown at the top) ClampR was performed on the full-length cDNA clone (lane 1 in each set) and on dsRNA isolated 3 weeks (lane 2 of each set) and 9 weeks (lane 3 of each set) posttransfection. Transfected hyphae were transferred from regeneration plates to PDA plates at 7 days posttransfection. After incubation for 7 days, the cultures were transferred to liquid medium for an additional week of culturing and then harvested for recovery of dsRNA (3-week samples) or transferred weekly to new PDA plates for 6 weeks (9-week samples) before transfer to liquid medium. Primer sets were chosen to amplify fragments spanning the foreign gene insert as follows: NS7 and BR54 (nucleotide map positions 476 to 493 and 1386 to 1402, respectively) for vectors p29Δ25-243EGFP, p29Δ25-243EGFP2A, p29Δ25-109EGFP, and p29Δ25-109EGFP2A; NS21 and NS22 (nucleotide map positions 2240 to 2259 and 2382 to 2401, respectively) for Ctp40[2A], Ct2APH, Ct2APHp29Δ25-243, and Ctp40[2AEGFP]; and BR16 and NS22 (nucleotide map positions 364 to 382 and 2382 to 2401, respectively) for Δp69bEGFP and Δp69bEGFP2A. Amplified fragments were electrophoresed in a 2.0% agarose gel in 1× Tris-borate-EDTA. M refers to the 100-bp DNA ladder size markers (Life Technologies). Arrows indicate the migration positions of the 2.1-, 0.6-, and 0.1-kbp size standards.