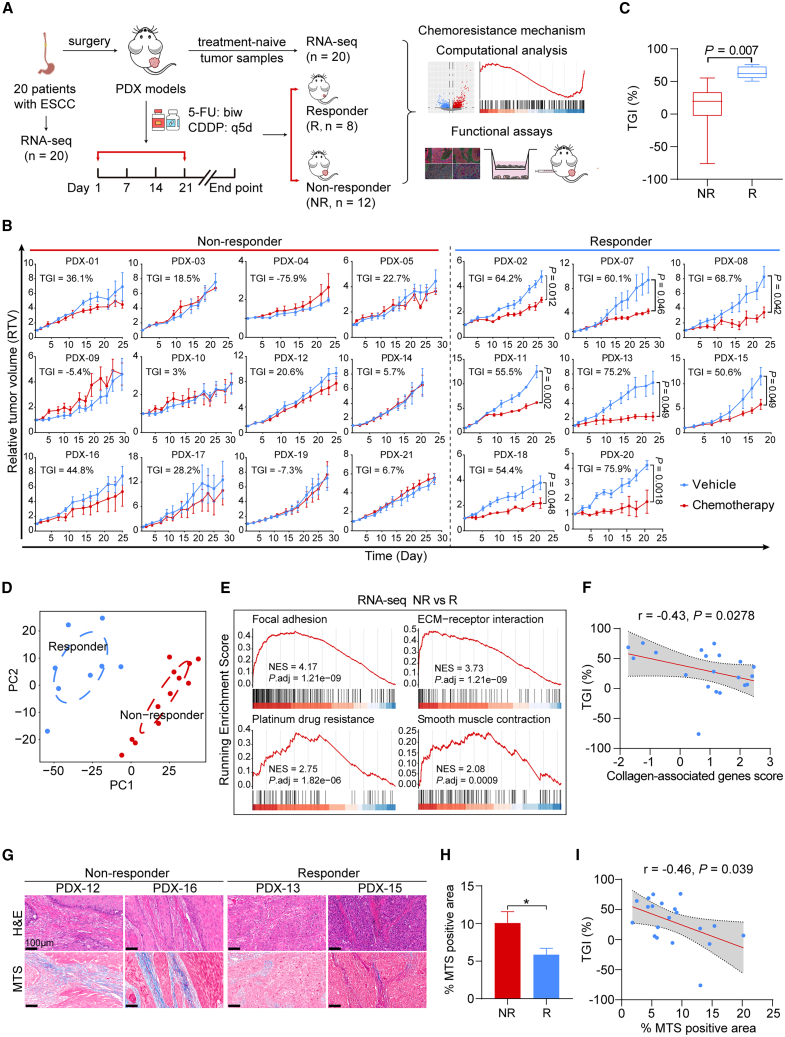
Figure 1.
Generation of ESCC PDXs chemotherapy cohort and identification of myCAF-associated gene signatures in non-responsive PDXs
(A) Graphical overview of the study.
(B) RTV curves of tumor xenografts from 20 distinct ESCC PDXs that were treated with chemotherapeutics or vehicle (n = 3–5 mice per group). Data are presented as mean ± SD. p values are determined using two-tailed Student’s t test.
(C) Boxplot exhibiting the percentage of tumor growth inhibition (TGI) of PDXs between the R (n = 8) and NR (n = 12) groups. Boxes represent the interquartile range, and whiskers represent the minimum and maximum values. p value is determined using two-tailed Student’s t test.
(D) Principal-component analysis (PCA) plot showing overall patterns of gene expression of 20 PDX donors’ tumor tissues. Circles indicate two separate clusters (R cluster [n = 8], blue; NR cluster [n = 12], red).
(E) Gene set enrichment analysis (GSEA) of pathways enriched in the NR group, using RNA-seq data of PDX donors’ tumor tissues.
(F) Spearman correlation between the TGI (%) and the collagen-associated genes score. The gray area represents 95% CI (n = 20).
(G) Representative images of H&E staining (top) and MTS (bottom) in the PDX donors’ biopsies of the R (n = 8) and NR (n = 12) groups. Scale bar, 100 μm.
(H) Quantification of (G). Data are presented as mean ± SEM. ∗p < 0.05 of two-tailed Student’s t test.
(I) Spearman correlation between the TGI (%) and the MTS-positive area (%). The gray area represents 95% CI (n = 20). See also Figure S1 and Table S1.