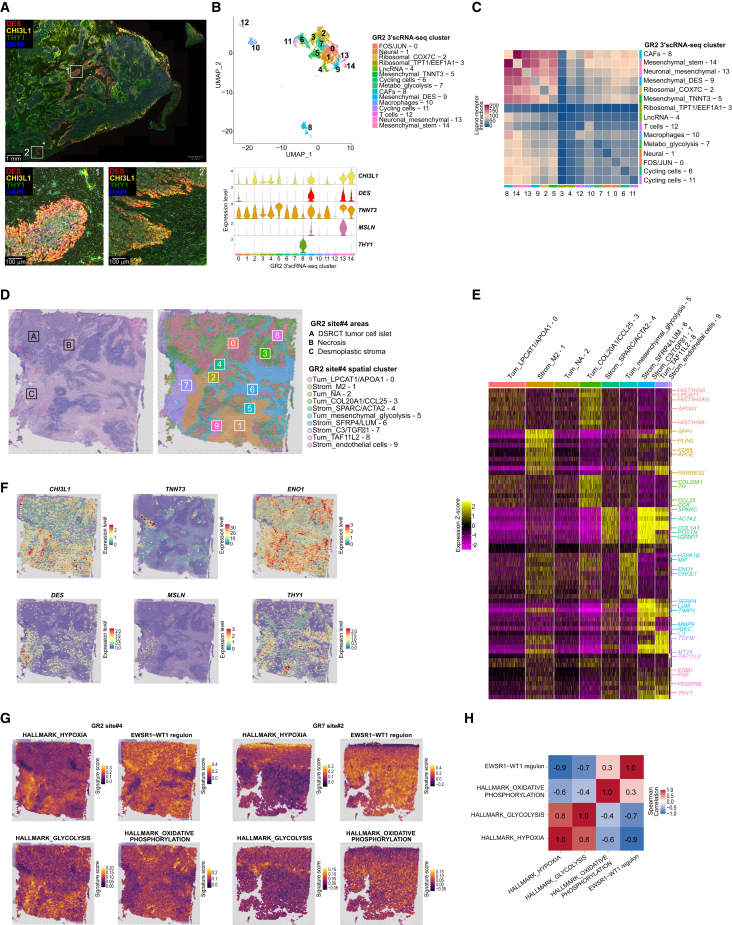
Figure 5.
DSRCT heterogeneity is linked to tumor spatial organization
(A) Immunofluorescent staining highlighting mesenchymal DSRCT tumor cells (DES+ and/or CHI3L1+), and CAFs (THY1+) on GR2 sample.
(B) UMAP showing GR2 3′ scRNA-seq clustering (upper panel), and violin plot (bottom panel) showing the expression of (1) tumor cell mesenchymal markers CHI3L1, DES, TNNT3, and MSLN; and (2) the CAF-specific marker THY1.
(C) Ligand-receptor interactions (CellPhoneDB) between cell clusters in the GR2 sample, representative of all specimens.
(D) GR2 site#4 sample annotated H&E-stained slide used for Visium assay, and spatial representation of spots’ clusters.
(E) Heatmap highlighting expression Z score of the top 10 DEG across spots’ clusters from GR2 site#4 Visium assay.
(F) Spatial representation of CHI3L1, TNNT3, ENO1, DES, MSLN and THY1 expression levels in GR2 site#4 sample using the Visium assay.
(G) Spatial gene signatures scores for HALLMARK_HYPOXIA, GLYCOLYSIS, OXPHOS, and EWSR1::WT1 regulon in GR2 site#4 and GR7 site#2 samples.
(H) Median Spearman correlation coefficients between gene signatures and EWSR1::WT1 regulon across Visium assays (n = 6).