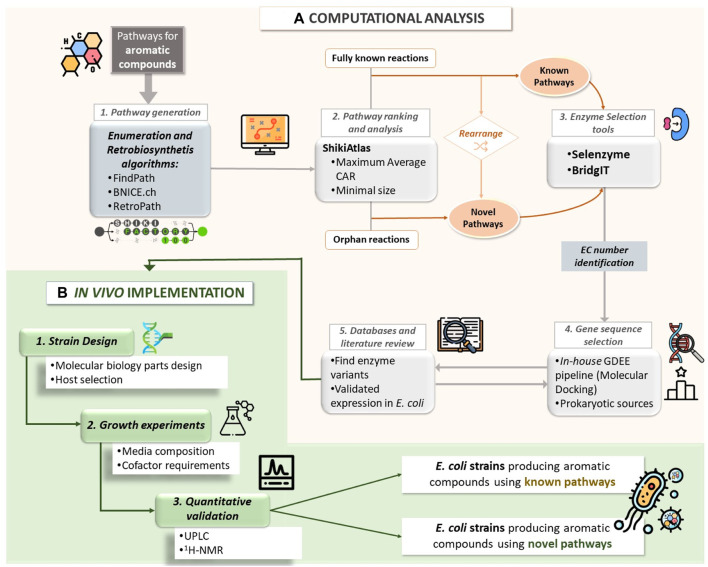
FIGURE 1.
Workflow of the (A) computational analysis of various biosynthetic pathways to yield aromatic compounds, including the generation of pathways through retrosynthesis and enumeration algorithms favoring maximum Conserved Atom Ratio (CAR) and minimal size. For the different reactions constituting the generated pathways, the enzyme selection tools Selenzyme and BrigIT were utilized to attribute an Enzyme Commission (EC) number. This step aided in the selection of the template enzyme for performing Molecular Docking using the in-house Gene Discovery and Enzyme Engineering (GDEE) pipeline. The enzymes were ranked based on their binding affinity score, and databases and literature were reviewed to finalize the selection of gene sequences. For the in vivo implementation (B), various host strains and molecular biology designs were tested. The cells were cultivated considering different media formulations, and the target compounds were quantified using Ultra Performance Liquid Chromatography (UPLC) and confirmed by 1H-NMR.