Abstract
We have determined the complete genome sequence of the Maastricht strain of rat cytomegalovirus (RCMV). The RCMV genome has a length of 229,896 bp and is arranged as a single unique sequence flanked by 504-bp terminal direct repeats. RCMV was found to have counterparts of all but one of the open reading frames (ORFs) that are conserved between murine CMV (MCMV) and human CMV (HCMV). Like HCMV, RCMV lacks homologs of the genes belonging to the MCMV m02 glycoprotein gene family. However, RCMV contains 15 ORFs with homology to members of the MCMV m145 glycoprotein gene family. Four ORFs are predicted to encode homologs of host proteins; R33 and R78 both putatively encode G protein-coupled receptors, whereas r144 and r131 encode homologs of major histocompatibility class I heavy chains and CC chemokines, respectively. An intriguing feature of the RCMV genome is the presence of an ORF, r127, with similarity to the rep gene of parvoviruses as well as ORF U94 of human herpesvirus 6A (HHV-6A) and HHV-6B. Counterparts of these ORFs have not been found in the other sequenced herpesviruses.
As a model for cytomegalovirus (CMV) infection and disease, we study the interaction between rat CMV (RCMV) and its host. The RCMV-rat model is attractive, since the pathogenesis of infection in RCMV-infected rats is similar to that in human CMV (HCMV)-infected humans. To fully exploit the RCMV-rat model, it is important to have a detailed picture of the genomic organization of RCMV. In this report, we present the complete DNA sequence of the RCMV (Maastricht) genome. Following HCMV (11) and murine CMV (MCMV) (37), RCMV is the third CMV for which the complete genome sequence has been determined. In addition, the RCMV genome represents the first complete sequence of a rat-specific herpesvirus.
General features of the RCMV genome sequence.
The RCMV genome was sequenced in a directed fashion, by using the previously cloned EcoRI and XbaI genomic subclones (32) as starting points. Overlapping plasmid clones of the genome were generated by using various restriction endonucleases. Both strands of each plasmid insert were sequenced by the dideoxynucleotide chain termination method. The final sequence was determined on both strands over 100% of the RCMV genome. Sequence assembly and analysis was done with the program PC/Gene (version 2.11; IntelliGenetics, Mountain View, Calif.) and with software from the United Kingdom human genome mapping project resource center (Hinxton, Cambridge, United Kingdom [http://www.hgmp.mrc.ac.uk]).
The length of the RCMV (Maastricht) genome was found to measure 229,896 bp, which is approximately 6 kb larger than a previous estimate that was based on an analysis of viral genomic restriction fragments (32). Remarkably, the genome of RCMV is only 382 bp shorter than that of MCMV (Smith) (37). The RCMV genome has an overall G+C content of 61% and consists of a single unique sequence flanked by 504-bp direct terminal repeats (TRs). The TRs are highly G+C rich (76%) and not represented elsewhere in the genome. Within the TRs, several small internal direct repeats (DRs) have been identified as well as conserved pac-1 and pac-2 sequences which are suspected to play a role in herpesvirus genome maturation (49). In addition, numerous other repeated sequences were identified throughout the RCMV genome, such as a variety of DRs and inverted repeats near the origin of lytic-phase DNA replication (50), and several DRs in the region upstream of exon 1 of the major immediate-early (MIE) locus (3).
Identification of RCMV protein-coding ORFs.
The strategy used to identify RCMV open reading frames (ORFs) likely to be coding was essentially based on that used in the sequence analysis of the MCMV genome (37). The major criteria for identifying a coding sequence were the presence of an ORF with a minimum length of 300 bp and a less than 60% overlap with adjacent ORFs. Obviously, the demonstration of similarity between predicted amino acid sequences encoded by RCMV ORFs and those encoded by well-characterized genes of other origin was also used as an indication of an ORF being protein coding. ORFs which were found to overlap more than 60% and not show any similarity to known sequences were included in the list of ORFs (Table 1), irrespective of their length and positional base preference. Database searches for homologous amino acid sequences were carried out with the TBLASTN program (version 2.0; National Center for Biotechnology Information, National Institutes of Health, Bethesda, Md. [http://www.ncbi.nlm.nih.gov/blast/blast.cgi?Jform=1]) against nonredundant combined nucleotide sequence databases. The TBLASTN program compares a protein query sequence against a nucleotide sequence database dynamically translated in all reading frames. The use of this program allows finding homologous amino acid sequences, even when these sequences are not available from protein sequence databases. The naming system used for RCMV ORFs numbers them from the left to the right end of the genome in a similar fashion as has been described for MCMV (37). The left-to-right orientation of the RCMV genome has previously been established (8, 50). As the RCMV and MCMV genomes were found to be largely colinear, the numbering system for the RCMV genes is congruent with the MCMV numbering system. RCMV ORFs with homologs in HCMV are indicated by uppercase prefixes (e.g., R23), whereas ORFs without significant sequence similarity with HCMV genes are indicated by lowercase prefixes. In order to maintain the correlation between the numbering system of the CMV genes, suffixes (as in r25.1) were introduced when additional unique RCMV ORFs were identified between homologs of MCMV and HCMV genes. These suffixes do not necessarily indicate any similarity between these RCMV ORFs. Also, these suffixes were used when RCMV ORFs showed similarity with MCMV ORFs with similar suffixes.
TABLE 1.
Map locations and features of the 166 predicted ORFs of the RCMV genomea
| ORF | Strandb | Positionc
|
Length (aa) | MM (kDa) | Identity (%) withd:
|
Comments (references)e | ||
|---|---|---|---|---|---|---|---|---|
| From | To | MCMV | HCMV | |||||
| r1 | C | 519 | 1172 | 218 | 25.5 | |||
| r2 | C | 1639 | 5358 | 1240 | 125.6 | |||
| r2.1 | 2912 | 4198 | 429 | 41.9 | ||||
| r3 | 5155 | 5814 | 220 | 24.5 | ||||
| r4 | C | 5753 | 6613 | 287 | 30.2 | |||
| r4.1 | 5787 | 6527 | 247 | 25.1 | ||||
| r5 | C | 6592 | 8478 | 629 | 65.1 | |||
| r5.1 | 7790 | 8113 | 108 | 11.8 | ||||
| r6 | 8477 | 8893 | 139 | 15.7 | ||||
| R23 | C | 8779 | 9663 | 295 | 31.9 | 41.2 (M23) | 18.9 (UL23 [GF2]) | US22 family homolog |
| r23.1 | 9662 | 10417 | 252 | 27.2 | ||||
| R24 | C | 10158 | 11195 | 346 | 36.9 | 46.3 (M24) | 27.6 (UL24 [GF2]) | US22 family homolog |
| R25 | 11333 | 13768 | 812 | 89.4 | 32.5 (M25) | 21.3 (UL25 [GF1]) | UL25 family homolog | |
| r25.1 | C | 13919 | 15079 | 387 | 42.5 | 29.2 (m25.1) | US22 family homolog | |
| r25.2 | 15878 | 16321 | 148 | 17.0 | ||||
| r25.3 | C | 15896 | 16258 | 121 | 13.2 | |||
| R26 | C | 16296 | 17027 | 244 | 25.9 | 36.5 (M26) | 29.0 (UL26) | |
| R27 | C | 17245 | 19257 | 671 | 76.1 | 39.6 (M27) | 20.7 (UL27) | |
| r27.1 | 19374 | 19745 | 124 | 13.3 | ||||
| R28 | C | 19720 | 20901 | 394 | 44.3 | 42.8 (M28) | 20.1 (UL28) | |
| r29.1 | C | 21135 | 21692 | 186 | 21.4 | 28.4 (m29.1) | ||
| R31 | 21691 | 24009 | 773 | 86.0 | 32.3 (M31) | 23.9 (UL31) | ||
| R32 | C | 24181 | 26181 | 667 | 73.0 | 37.4 (M32) | 18.2 (UL32 [pp150]) | Homolog of HCMV gene encoding major tegument phosphoprotein (7) |
| R33 | 26271 | 27431 | 387 | 43.2 | 64.2 (M33) | 37.3 (UL33) | GPCR gene homolog (5) | |
| R34 | 27693 | 29990 | 766 | 84.6 | 38.9 (M34) | 22.7 (UL34) | ||
| R35 | 30318 | 31880 | 521 | 58.2 | 45.8 (M35) | 24.3 (UL35 [GF1]) | UL25 family homolog | |
| R36 | C | 32054 | 33487 | 478 | 53.6 | 47.1 (M36) | 26.0 (UL36) | US22 family homolog |
| R37 | C | 33708 | 34700 | 331 | 36.5 | 37.2 (M37) | 17.3 (UL37) | |
| R38 | C | 34818 | 35933 | 372 | 41.0 | 32.9 (M38) | 25.3 (UL38) | |
| r39 | C | 36407 | 37012 | 202 | 22.3 | 26.1 (m39) | ||
| r40 | C | 37079 | 37471 | 131 | 14.4 | 39.4 (m40) | ||
| r41 | C | 37619 | 38035 | 139 | 14.5 | 33.3 (m41) | ||
| r42 | C | 38124 | 38504 | 127 | 13.5 | 27.6 (m42) | ||
| R43 | C | 38825 | 40504 | 560 | 61.8 | 40.3 (M43) | US22 family homolog; positional homolog of HCMV UL43 | |
| r43.1 | C | 40582 | 40896 | 105 | 11.1 | |||
| R44 | C | 41045 | 42301 | 419 | 45.6 | 68.9 (M44) | 55.2 (UL44 [DPAP]) | Homolog of HCMV gene encoding DPAP |
| R45 | C | 42687 | 45878 | 1064 | 114.2 | 33.8 (M45) | 25.3 (UL45 [RRL]) | Homolog of HCMV gene encoding RRL |
| R46 | C | 45895 | 46776 | 294 | 33.1 | 73.8 (M46) | 35.8 (UL46) | Homolog of HCMV gene encoding minor capsid protein |
| R47 | 46775 | 49675 | 967 | 109.0 | 55.8 (M47) | 28.9 (UL47) | ||
| R48 | 49675 | 55974 | 2100 | 234.5 | 51.8 (M48) | 27.0 (UL48 [Teg]) | Homolog of HCMV gene encoding large tegument (Teg) protein | |
| R49 | C | 56351 | 57925 | 525 | 59.3 | 71.3 (M49) | 37.6 (UL49) | |
| R50 | C | 57900 | 58787 | 296 | 32.6 | 53.5 (M50) | 36.7 (UL50) | |
| R51 | C | 58814 | 59197 | 128 | 14.2 | 69.5 (M51) | 39.5 (UL51) | |
| R52 | 59208 | 60797 | 530 | 60.5 | 66.9 (M52) | 35.5 (UL52) | ||
| R53 | 60793 | 61698 | 302 | 35.3 | 62.8 (M53) | 36.6 (UL53) | ||
| R54 | C | 61692 | 65210 | 1173 | 130.5 | 57.3 (M54) | 43.8 (UL54 [DNA pol]) | Homolog of HCMV gene encoding DNA polymerase (DNA pol) (8) |
| R55 | C | 65223 | 67964 | 914 | 102.7 | 53.8 (M55) | 43.2 (UL55 [gB]) | Homolog of HCMV gene encoding glycoprotein B (gB) (8) |
| R56 | C | 67873 | 70626 | 893 | 97.2 | 66.7 (M56) | 50.2 (UL56) | Homolog of HCMV gene encoding ICP18.5 (8) |
| R57 | C | 70828 | 74670 | 1281 | 140.0 | 51.9 (M57) | 45.2 (UL57 [MDBP]) | Homolog of HCMV gene encoding major DNA-binding protein (MDBP) (8) |
| r58 | 74474 | 75337 | 288 | 30.1 | 27.3 (m58) | |||
| R69 | C | 79600 | 82593 | 998 | 109.4 | 35.0 (M69) | 22.5 (UL69) | Homolog of HCMV gene encoding a transactivator of gene expression |
| R70 | C | 82820 | 85627 | 936 | 105.8 | 50.3 (M70) | 37.9 (UL70 [HP]) | Homolog of HCMV gene encoding a helicase-primase (HP) complex component |
| r70.1 | 86170 | 86484 | 105 | 11.0 | ||||
| r70.2 | 86689 | 87681 | 331 | 36.3 | Member of the m145 gene family | |||
| r70.3 | 87847 | 88872 | 342 | 37.0 | Member of the m145 gene family | |||
| r70.4 | 89015 | 90034 | 340 | 36.7 | Member of the m145 gene family | |||
| r70.5 | 90158 | 91171 | 338 | 36.8 | Member of the m145 gene family | |||
| R72 | C | 91238 | 92278 | 347 | 38.5 | 35.6 (M72) | 24.1 (UL72 [dUTPase]) | Homolog of HCMV gene encoding dUTPase |
| R73 | 92277 | 92648 | 124 | 13.9 | 48.3 (M73) | 28.1 (UL73) | ||
| r74 | C | 92526 | 93941 | 472 | 53.9 | 23.0 (m74) | ||
| R75 | C | 94262 | 96469 | 736 | 81.7 | 44.7 (M75) | 26.8 (UL75 [gH]) | Homolog of HCMV gene encoding glycoprotein H (gH) |
| R76 | 96590 | 97360 | 257 | 28.4 | 53.3 (M76) | 36.3 (UL76) | ||
| R77 | 96996 | 98963 | 656 | 71.3 | 53.4 (M77) | 40.8 (UL77) | Homolog of HCMV gene encoding pyruvoyl decarboxylase homolog | |
| R78 | 99095 | 100516 | 474 | 49.6 | 25.0 (M78) | 20.1 (UL78) | GPCR gene homolog (2) | |
| R79 | C | 100737 | 101558 | 274 | 31.3 | 67.9 (M79) | 41.4 (UL79) | |
| R80 | 101557 | 103743 | 729 | 75.9 | 46.0 (M80) | 33.0 (UL80 [AP]) | Homolog of HCMV gene encoding assembly protein (AP) | |
| R82 | C | 104532 | 106334 | 601 | 66.6 | 33.2 (M82) | 23.5 (UL82 [pp71]) | Homolog of HCMV gene encoding upper matrix phosphoprotein |
| R83 | C | 106489 | 108429 | 647 | 71.6 | 28.1 (M83) | 19.2 (UL83 [pp65]) | Homolog of HCMV gene encoding lower matrix phosphoprotein |
| R84 | C | 108562 | 110361 | 600 | 65.9 | 33.6 (M84) | 20.2 (UL84) | Homolog of HCMV gene encoding an early nuclear nonstructural protein |
| R85 | C | 110473 | 111396 | 308 | 34.0 | 68.6 (M85) | 52.7 (UL85) | |
| R86 | C | 111501 | 115547 | 1349 | 150.9 | 77.3 (M86) | 56.8 (UL86 [MCP]) | Homolog of HCMV gene encoding major capsid protein (MCP) |
| R87 | 115605 | 118244 | 880 | 97.2 | 72.0 (M87) | 46.3 (UL87) | ||
| R88 | 118278 | 119582 | 435 | 47.6 | 54.5 (M88) | 28.1 (UL88) | ||
| R89-EX2 | C | 119585 | 120706 | 374 | 42.5 | 89.0 (M89-EX2) | 69.8 (UL89 [CHS]) | Homolog of exon 2 of HCMV gene encoding conserved herpesvirus spliced gene (CHS); exon 1 plus exon 2 encode a protein of 670 amino acids with an MM of 77.1 kDa (Y. K. Gruijthuijsen, C. A. Bruggeman, and C. Vink, unpublished data) |
| r90 | C | 120861 | 121646 | 262 | 27.7 | 21.1 (m90) | ||
| R91 | 121360 | 122079 | 240 | 24.9 | 33.3 (M91) | 15.8 (UL91) | ||
| R92 | 122079 | 122792 | 238 | 26.2 | 81.2 (M92) | 45.0 (UL92) | ||
| R93 | 122758 | 124281 | 508 | 56.3 | 51.1 (M93) | 27.9 (UL93) | ||
| R94 | 124241 | 125269 | 343 | 37.1 | 51.4 (M94) | 33.0 (UL94) | ||
| R89-EX1 | C | 125423 | 126310 | 296 | 34.6 | 78.8 (M89-EX1) | 57.4 (UL89 [CHS]) | Homolog of exon 1 of HCMV gene encoding CHS |
| R95 | 126309 | 127484 | 392 | 42.7 | 65.5 (M95) | 41.3 (UL95) | ||
| R96 | 127577 | 127885 | 103 | 11.5 | 42.0 (M96) | 33.0 (UL96) | ||
| R97 | 128104 | 129939 | 612 | 66.9 | 53.1 (M97) | 32.8 (UL97 [PK]) | Homolog of HCMV gene encoding a phosphotransferase or protein kinase (PK) | |
| R98 | 130193 | 131665 | 491 | 53.6 | 47.3 (M98) | 36.3 (UL98 [DNase]) | Homolog of HCMV gene encoding an exonuclease | |
| R99 | 131605 | 131976 | 124 | 13.1 | 39.5 (M99) | 23.2 (UL99 [pp28]) | Homolog of HCMV gene encoding a tegument phosphoprotein | |
| R100 | C | 132208 | 133260 | 351 | 39.3 | 68.9 (M100) | 42.7 (UL100 [gM]) | Homolog of HCMV gene encoding glycoprotein M (gM) |
| R102 | 133471 | 136428 | 986 | 106.7 | 29.5 (M102) | 23.8 (UL102 [HP]) | Homolog of HCMV gene encoding a HP complex component | |
| R103 | C | 135996 | 137126 | 377 | 41.1 | 44.4 (M103) | 24.5 (UL103) | |
| R104 | C | 137107 | 139317 | 737 | 83.1 | 63.7 (M104) | 39.9 (UL104) | Homolog of HCMV gene encoding a structural protein |
| R105 | 139118 | 141949 | 944 | 104.8 | 58.5 (M105) | 48.7 (UL105 [Hel]) | Homolog of HCMV gene encoding DNA helicase (Hel) | |
| r106 | C | 142119 | 142610 | 164 | 18.0 | |||
| r107 | C | 147501 | 147884 | 128 | 14.2 | |||
| r108 | 147846 | 148187 | 114 | 12.7 | ||||
| r109 | C | 149116 | 149445 | 110 | 12.5 | |||
| r110 | C | 149808 | 150116 | 103 | 118.8 | |||
| r111.1 | 152167 | 152508 | 114 | 12.3 | ||||
| r111.2 | C | 152241 | 152549 | 103 | 11.0 | |||
| R112-EX1 | 153431 | 154184 | 251 | 26.7 | 50.9 (M112-EX1) | 29.2 (UL112) | ||
| R112-EX2 | 154408 | 154632 | 74 | 7.5 | 50.6 (M112-EX2) | 26.3 (UL112) | ||
| R113 | 154665 | 155699 | 345 | 35.4 | 29.7 (M113) | 25.8 (UL113) | ||
| R114 | C | 155910 | 156686 | 259 | 29.6 | 68.1 (M114) | 50.2 (UL114 [UNG]) | Homolog of HCMV gene encoding uracil DNA glycosylase (UNG) |
| R115 | C | 156745 | 157689 | 315 | 35.9 | 44.4 (M115) | 27.2 (UL115 [gL]) | Homolog of HCMV gene encoding glycoprotein L (gL) |
| R116 | C | 157774 | 158982 | 403 | 45.2 | 16.0 (M116) | 18.0 (UL116) | |
| R117 | C | 159067 | 160194 | 376 | 41.6 | 25.9 (m117) | 18.7 (UL117) | |
| R118 | C | 160263 | 161111 | 283 | 32.6 | 27.0 (M118) | 19.0 (UL118) | |
| r119.1 | C | 161175 | 161903 | 243 | 27.9 | 28.1 (m119.1) | ||
| r119.2 | C | 162094 | 162432 | 113 | 12.6 | 34.1 (m119.2) | ||
| r119.3 | C | 162442 | 162759 | 106 | 11.3 | 25.7 (m119.3) | ||
| r119.4 | C | 163546 | 164562 | 339 | 37.1 | |||
| r119.5 | C | 164713 | 165699 | 329 | 36.5 | |||
| r119.6 | C | 165836 | 166756 | 307 | 34.1 | 26.2 (novel) | Homolog of a novel MCMV ORF, located at position 174640 to 175665 of the MCMV genome, putatively encoding a 342-amino-acid protein | |
| R121 | C | 167155 | 168753 | 533 | 61.0 | 18.8 (M121) | Positional homolog of HCMV UL121 | |
| r121.1 | 169295 | 169666 | 124 | 12.7 | ||||
| r121.2 | C | 169302 | 169715 | 138 | 15.2 | |||
| r121.3 | C | 169938 | 170237 | 100 | 11.6 | |||
| R122-EX5 | C | 170236 | 171749 | 505 | 56.8 | 37.3 (M122-EX5) | 26.3 (UL122 [IE2]) | Exon 5 of MIE 2 (IE2) gene; homolog of HCMV IE2 and MCMV ie3; exon 2 plus exon 3 plus exon 5 encode a protein of 603 amino acids with an MM of 67.8 kDa (3) |
| r123-EX4 | C | 171961 | 173252 | 431 | 49.8 | 20.0 (M123-EX4) | 15.3 (UL123 [IE1]) | Exon 4 of MIE 1 (IE1) gene; homolog of HCMV IE1 and MCMV ie1; exon 2 plus exon 3 plus exon 4 encode a protein of 529 amino acids with an MM of 60.8 kDa (3) |
| r123-EX3 | C | 173349 | 173542 | 64 | 7.0 | 37.3 (m123-EX3) | Exon 3 of MIE locus (3) | |
| r123-EX2 | C | 173640 | 173740 | 34 | 4.0 | 24.3 (m123-EX2) | Exon 2 of MIE locus (3) | |
| r124 | 173688 | 174035 | 116 | 13.6 | 17.6 (m124) | |||
| r125 | 175431 | 175751 | 107 | 11.8 | ||||
| r126 | C | 177276 | 177578 | 101 | 11.6 | |||
| r127 | C | 178309 | 179319 | 337 | 37.8 | Homolog of parvovirus rep gene and HHV-6 U94 gene | ||
| r128 | 179479 | 180702 | 408 | 47.0 | 42.9 (m128) | US22 family homolog; homolog of MCMV ie2 exon 3 | ||
| r131 | C | 181948 | 182649 | 234 | 26.8 | 22.5 (m131/129) | Homolog of spliced MCMV m131/129 transcript encoding a CC chemokine homolog | |
| r133 | C | 183022 | 184110 | 363 | 40.6 | 26.1 (m133-EX1) | Homolog of exon 1 of spliced MCMV gene m133/132 (sgg1) | |
| r135 | C | 184218 | 184592 | 125 | 13.5 | 33.3 (m135) | ||
| r136 | C | 184669 | 185430 | 254 | 29.1 | 35.1 (m136) | ||
| r137 | C | 185448 | 186401 | 318 | 34.9 | 35.5 (m137) | ||
| r138 | C | 186464 | 188149 | 562 | 62.7 | 24.3 (m138) | Homolog of MCMV gene encoding an Fc receptor glycoprotein | |
| r139 | C | 188287 | 190251 | 655 | 73.6 | 43.5 (m139) | 21.6 (US22 [GF2]) | US22 family homolog |
| r140 | C | 190311 | 191765 | 485 | 55.5 | 45.4 (m140) | 29.5 (US23 [GF2]) | US22 family homolog |
| r141 | C | 191892 | 193382 | 497 | 56.1 | 45.3 (m141) | 26.2 (US24 [GF2]) | US22 family homolog |
| r142 | C | 193609 | 195255 | 549 | 62.0 | 57.3 (m142) | 21.0 (US26 [GF2]) | US22 family homolog |
| r143 | C | 195104 | 196693 | 530 | 60.0 | 43.2 (m143) | 23.2 (US23 [GF2]) | US22 family homolog |
| r144 | C | 196862 | 197824 | 321 | 36.2 | 30.4 (m144) | MHC class I gene homolog (4) | |
| r145 | C | 198005 | 199351 | 449 | 50.8 | 24.6 (m145) | Member of the m145 gene family | |
| r146 | C | 199486 | 199857 | 124 | 14.2 | |||
| r147 | C | 200217 | 201083 | 289 | 32.7 | |||
| r148 | C | 201125 | 201499 | 125 | 14.6 | |||
| r149 | C | 201629 | 202768 | 380 | 44.4 | Member of the m145 gene family | ||
| r150 | C | 202926 | 204095 | 390 | 43.4 | 20.1 (m150) | Member of the m145 gene family | |
| r151 | C | 204273 | 205385 | 371 | 41.5 | 22.8 (m151) | Member of the m145 gene family | |
| r151.1 | C | 205520 | 206395 | 292 | 33.4 | |||
| r151.2 | C | 206562 | 207497 | 312 | 35.9 | |||
| r151.3 | C | 207686 | 209377 | 564 | 62.7 | Member of the m145 gene family | ||
| r152 | C | 209879 | 211030 | 384 | 43.8 | 19.0 (m152) | Member of the m145 gene family | |
| r152.1 | C | 211471 | 211914 | 148 | 17.2 | |||
| r152.2 | C | 212063 | 213130 | 356 | 41.0 | Member of the m145 gene family | ||
| r152.3 | C | 213310 | 214209 | 300 | 35.0 | Member of the m145 gene family | ||
| r152.4 | C | 214454 | 215590 | 379 | 43.4 | Member of the m145 gene family | ||
| r152.5 | C | 215829 | 216611 | 261 | 30.5 | |||
| r155 | C | 217788 | 218804 | 339 | 39.1 | 20.2 (m155) | Member of the m145 gene family | |
| r157 | C | 218951 | 220030 | 360 | 42.0 | 19.7 (m157) | Member of the m145 gene family | |
| r158 | C | 220209 | 220544 | 112 | 12.5 | |||
| r160 | C | 220586 | 221377 | 264 | 29.6 | 23.5 (m160) | Homolog of putative membrane glycoprotein gene of MCMV | |
| r161 | C | 221232 | 222242 | 337 | 37.7 | 18.2 (m161) | Homolog of putative membrane glycoprotein gene of MCMV | |
| r162 | C | 222146 | 223036 | 297 | 32.2 | 20.3 (m162) | Homolog of putative membrane glycoprotein gene of MCMV | |
| r164 | C | 223119 | 224261 | 381 | 42.6 | 22.6 (m164) | Homolog of putative membrane glycoprotein gene of MCMV | |
| r166 | C | 225087 | 226208 | 374 | 41.1 | 27.6 (m166) | Homolog of putative membrane glycoprotein gene of MCMV | |
| r167 | C | 226211 | 226831 | 207 | 22.9 | |||
| r168 | C | 226983 | 227297 | 105 | 12.1 | |||
| r169 | C | 227397 | 228038 | 214 | 23.2 | |||
| r170 | 227961 | 228281 | 107 | 12.4 | ||||
| r171 | 228242 | 229105 | 288 | 32.6 | ||||
| r171.1 | 228439 | 229101 | 221 | 24.4 | ||||
Abbreviations and symbols: aa, number of amino acids; MM, molecular mass; EX1-5, exons 1 to 5 of (potentially) spliced genes.
C indicates that the corresponding ORF runs from right to left on the prototype RCMV genome (see Fig. 1); other ORFs run from left to right.
From and To indicate the limits of the ORFs on the prototype RCMV genome.
In columns MCMV and HCMV, the percentages of identity are shown when significant similarity had been determined; the names of MCMV (Smith) ORFs (37) and HCMV (AD169) ORFs (11) that show similarity with RCMV ORFs are shown in brackets. The percentages of identity were determined by using the ALIGN program, which generates optimal global alignments of two sequences with no short-cuts (Genestream, Institut de Genetique Humaine, Montpellier, France [http://www2.igh.cnrs.fr/bin/align-guess.cgi]) (35). In order to generate optimal alignments, the following ORFs were trimmed to a downstream ATG codon: HCMV UL70 and UL95, MCMV M51, M96, M100 and m119.1, and RCMV M115 and m142.
Characteristics of the ORF, including references to previous studies on this ORF.
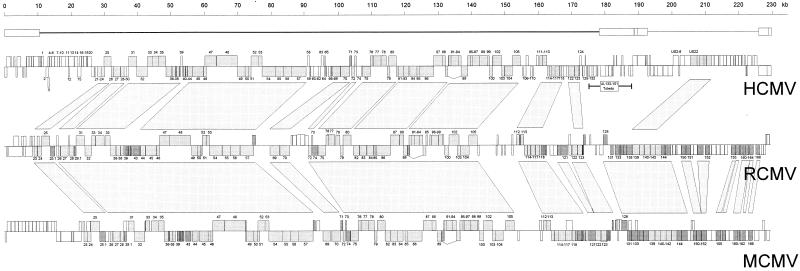
The RCMV genome is predicted to contain at least 166 protein-coding ORFs, of which 113 and 76 have significant similarity to ORFs of MCMV (Smith) (37) and HCMV (AD169) (11), respectively (Fig. 1 and Table 1). ORFs that are conserved between RCMV and HCMV are concentrated within the left two-thirds of the HCMV genome (Fig. 2). However, as expected, the similarity between RCMV and MCMV ORFs is seen across the entire length of their genomes. All ORFs that are conserved among members of the herpesvirus family also have counterparts in the RCMV genome. For all conserved RCMV ORFs, the degree of similarity gradually decreases with corresponding sequences of MCMV, HCMV (Table 1), and other betaherpesviruses (human herpesvirus 6A [HHV-6A], HHV-6B, and HHV-7; data not shown). The highest level of identity was seen among RCMV R89, MCMV M89, and HCMV UL89.
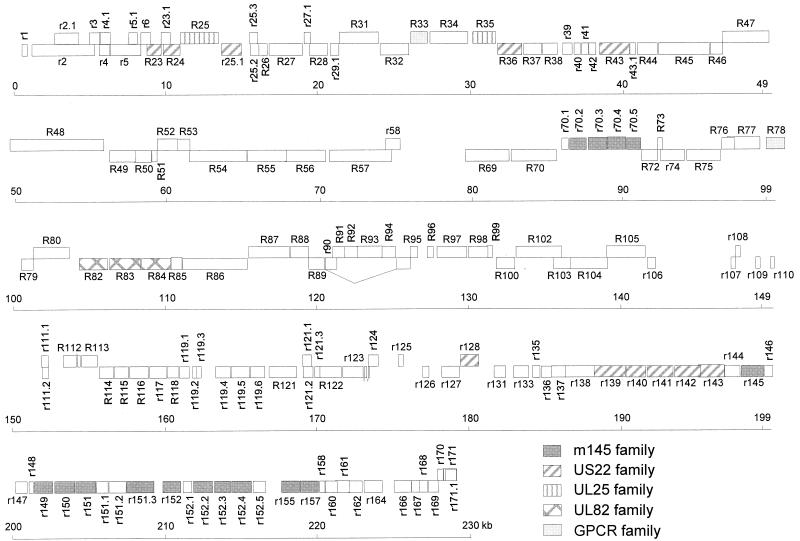
FIG. 1.
Map of the 229,896-kb RCMV (Maastricht) genome. ORFs are shown as boxes. ORFs on the top strand (coding left to right) are shown above those on the bottom strand (coding right to left). The ORFs are numbered as described in the text and are defined as indicated in Table 1. Members of the five RCMV gene families, the m145, US22, UL25, UL82, and GPCR families, are indicated. The exons of RCMV ORFs that are either known or predicted to encode spliced transcripts, R89 (Y. K. Gruijthuijsen, C. A. Bruggeman, and C. Vink, unpublished data), R112, and R122-123 (3), are connected by lines.
FIG. 2.
Alignment of the ORF maps of the genomes of RCMV, HCMV, and MCMV. The line above the HCMV map indicates the UL-US organization of the HCMV genome. For each genome map, the ORFs (boxes) on the top strand (coding left to right) are shown above those on the bottom strand (coding right to left). Boxes that represent ORFs that are conserved between HCMV, RCMV, and MCMV are depicted in light grey; boxes that represent ORFs that are conserved between only two of the three viruses are shown in dark grey. The numbers of most ORFs (without their prefixes) are indicated. ORFs that are conserved between RCMV and the other two CMVs are connected by blocks. The MCMV map is based on Rawlinson et al. (37), and the HCMV map is based on Chee et al. (11) and Cha et al. (9).
As in the MCMV genome, homologs of the HCMV gene families UL25, UL82, and US22 are present in the RCMV sequence, whereas counterparts of the RL11, US1, US2, US6, or US12 gene families of HCMV are absent. Previously, HCMV strains Toledo and Towne were shown to have 19 and 3 extra ORFs, respectively, in addition to those found in HCMV (AD169) (9). Homologs of these additional ORFs were not found in the RCMV genome. Unlike HCMV, RCMV possesses homologs of genes belonging to the MCMV m145 glycoprotein gene family. Fifteen RCMV members of this family were identified, most of which are located at positions within the genome that are congruent to the positions of their MCMV counterparts, near the right genome terminus.
A striking difference between MCMV and RCMV is seen at the left side of their genomes. Within the MCMV sequence, a series of 15 tandem glycoprotein genes (the m02 glycoprotein gene family) is positioned between nucleotides 999 and 15673 (37). Homologs of these genes were found neither in the sequence of RCMV nor in that of HCMV.
Spliced transcripts.
Splicing has previously been reported for RCMV R89 (Y. K. Gruijthuijsen, C. A. Bruggeman, and C. Vink, unpublished data) and R122/R123 (MIE) (3). Splicing of two other RCMV transcripts, R112 and r133, could be predicted on the basis of amino acid sequence alignments and the presence of consensus splice donor and acceptor sites (data not shown). In contrast to what was predicted for the MCMV M36 mRNA, transcripts from its RCMV homolog (R36) are probably not spliced. This notion was deduced from an alignment of the amino acid sequence derived from the unspliced R36 ORF with that derived from the spliced M36 ORF (data not shown). Similarly, transcripts from both the MCMV M33 and HCMV UL33 genes were reported to be spliced (16), whereas transcripts from their RCMV counterpart (R33) were demonstrated to be unspliced (5).
DNA sequences and proteins involved in nucleotide and DNA metabolism and DNA replication.
The origin of lytic-phase DNA replication (oriLyt) of RCMV has previously been mapped to a 3.3-kb region between ORFs R57 and R69 and was shown to be highly complex, containing 23 DRs and 16 inverted repeats of lengths greater than 10 bp (50). Like MCMV and HCMV, RCMV contains six ORFs that may be essential for viral DNA replication. These ORFs, R44, R54, R57, R70, R102, and R105, have the potential to code for DNA polymerase accessory protein (DPAP), DNA polymerase (8), major DNA binding protein (8), and three components of the helicase-primase complex, respectively. Homologs of genes with a role in nucleotide metabolism are also found in the RCMV genome. These genes include ORFs R45, R72, and R114, which putatively encode the ribonucleotide reductase large subunit (RRL), dUTPase, and uracil-DNA glycosylase, respectively. ORF R97 is the homolog of HCMV UL97, which encodes a phosphotransferase (28).
ORFs encoding IE/regulatory proteins.
Previously, several HCMV immediate early (IE) proteins were reported to play a role in the regulation of viral gene expression. These proteins are encoded by the UL122 to -123 (the MIE locus) (43), UL36 to -38 (27), UL69 (51), TRS1-IRS1 (42), and US3 (13) genes. RCMV possesses sequence and positional homologs of the first three loci (R122-123, R36-38, and R69). The organization of the R122-123 MIE locus (3) was previously shown to be similar to that of HCMV (43), MCMV (25), simian CMV (10), and the England strain of RCMV (39, 40). Various spliced transcripts are derived from each of these loci. Similarly, the HCMV UL36 to -38 IE locus was found to be transcribed in multiply spliced mRNAs. Some of the proteins that are encoded by these mRNAs have been reported to function in the activation of gene transcription (13, 24). One of the spliced transcripts from the UL36 to -38 gene cluster is the UL37 mRNA, which is composed of three exons. Only exon 3 of UL37 has homologs in the other sequenced betaherpesviruses.
Another potential IE gene of RCMV is r128, which is positioned several kilobases upstream of the MIE locus. ORF r128 is the homolog of the MCMV m128 (or ie2) IE gene (33) and has sequence and positional homology with the U95 ORFs of HHV-6A (20), HHV-6B (17, 22), and HHV-7 (36). ORF m128 was previously shown to have sequence similarity with members of the US22 gene family (33).
Structural proteins.
Homologs of all MCMV genes that have the capacity to encode the well-known structural proteins were also detected in the RCMV genome. These genes are likely to encode the major and minor capsid proteins (R86 and R46, respectively), the large tegument protein (R48), the upper (R82) and lower (R83) matrix proteins, and the major and small tegument phosphoproteins (R32 and R99, respectively) (10). In addition, the RCMV genome carries a homolog (R25) of MCMV M25, which was recently reported to code for a component of the viral tegument (52). Like their MCMV counterparts (14), RCMV ORFs R82, R83, and R84 were found to share sequences. The overall identities among the amino acid sequences that are deduced from these ORFs are 21.2% between R82 and R83, 22.4% between R82 and R84, and 19.1% between R83 and R84. When the amino acid sequences encoded by R82, R83, and R84 were compared to their MCMV counterparts, the highest similarities were seen between sequences derived from congruent positions within their respective genomes. Interestingly, when the amino acid sequences deduced from R82, R83, and R84 were compared to those from their HCMV counterparts (UL82, UL83, and UL84, respectively), the three RCMV sequences each showed a higher similarity with sequences derived from UL82 than with those from either UL83 or UL84. Conversely, the three HCMV sequences each displayed higher similarities with the amino acid sequence encoded by R82 than with the sequences deduced from the other two RCMV genes (data not shown).
Glycoproteins.
Among the ORFs potentially encoding glycoproteins are R55 (8), R75, R100, and R115, which code for homologs of the conserved herpesvirus glycoproteins gB, gH, gM, and gL, respectively. In particular, the sequence of the putative RCMV gM protein is very similar to the sequence of the corresponding MCMV protein (68.9% identity). A glycoprotein may also be encoded by ORF r138, which is a homolog of the MCMV m138 gene (or fcr-1) (44). Homologs of this gene have not been identified in other betaherpesviruses. The m138 gene-encoded protein has been reported to be a receptor for the Fc domain of murine immunoglobulin G molecules (44). Recombinant MCMV strains that lack a functional m138 gene displayed severely restricted replication in comparison with wild-type MCMV in vivo (15).
Families of related RCMV ORFs.
Five families of related genes were identified in the RCMV genome: (i) the UL25 family, including ORF R25 and R35; (ii) the UL82 family, including R82, R83, and R84; (iii) the US22 family; (iv) the m145 family; and (v) the G protein-coupled receptor (GPCR) homolog gene family, including R33 and R78 (Fig. 1). These families are also represented in the MCMV genome (37), whereas all but one of them (the m145 family) are represented in the HCMV sequence (11).
Members of the US22 gene family are present in all sequenced betaherpesviruses. The RCMV members of this family include R23, R24, r25.1, R36, R43, r128, r139, r140, r141, r142, and r143. The sequence as well as the position of these genes are conserved between RCMV and MCMV. Within the RCMV US22 family, the highest level of similarity is seen between the amino acid sequences derived from R24 and r25.1 (25.8% identity). A relatively high amino acid sequence similarity is also observed between r140 and r141 (25.6% identity) and between r140 and r143 (22.9% identity). An RCMV homolog of one MCMV member of the US22 family, m25.2, was not found.
As described above, RCMV contains 15 members of the m145 glycoprotein gene family (Fig. 1). One of the members of this family, m152, has been shown to interfere with the major histocompatibility complex (MHC) class I pathway of antigen presentation (54). Six of the RCMV m145-like ORFs (r145, r150, r151, r152, r155, and r157) have both positional and sequence homology to the corresponding MCMV ORFs (37). Five others (r149, r151.3, r152.2, r152.3, and r152.4) show similarity in sequence, but not position, with MCMV ORFs. ORF r149 displays highest similarity with MCMV m17, whereas r151.3 scores highest with m145. ORFs r152.2, r152.3, and r152.4 are more similar to m152 than to other MCMV m145-like genes (data not shown). Four m145 family members, ORFs r70.2 through r70.5, are located at a unique position within the RCMV sequence, between conserved ORFs R70 and R72. Unlike their counterparts at the right side of the prototype genome, these ORFs are orientated from left to right. ORFs r70.2 to r70.5 were found to have the highest level of sequence similarity among each other. In particular, the deduced amino acid sequences of r70.2 and r70.4 are highly related (44.1% identity). ORFs r70.2 to r70.5 were also found to show a relatively high degree of similarity with ORF r152.2 (data not shown).
ORFs encoding homologs of cellular proteins.
Similar to MCMV, RCMV contains four ORFs that encode homologs of cellular proteins. R33 and R78 both encode homologs of GPCRs (2, 5), whereas r131 and r144 (4) encode homologs of chemokines and MHC class I molecules, respectively.
(i) ORFs encoding GPCR homologs.
RCMV ORF R33 belongs to the HCMV UL33 gene family (5, 48). Currently, this family consists of six members: UL33 (12), R33 (5), MCMV M33 (16), and the U12 ORFs of HHV-6A (20), HHV-6B (17, 22), and HHV-7 (36). Sequence and genome location of these genes are conserved among the betaherpesviruses. The predicted amino acid sequences of the proteins encoded by members of the UL33-like gene family were found to comprise several features characteristic of chemokine receptors (5, 16). In accordance with this, the HHV-6 U12-encoded protein was reported to be a functional receptor for β-chemokines in vitro (23). It has been shown that the UL33, M33, and R33 genes are dispensable for in vitro replication of HCMV (31), MCMV (16), and RCMV (5), respectively. However, both M33 and R33 were shown to be essential for in vivo replication of MCMV (16) and RCMV (5), respectively.
RCMV R78 belongs to the HCMV UL78 gene family, which currently consists of six members: UL78 (12), R78 (2), MCMV M78 (37), and the U51 ORFs of HHV-6A (20), HHV-6B (17, 22), and HHV-7 (36). Although the positions of the UL78-like genes within the betaherpesvirus genomes are conserved, their sequences are rather divergent (2, 48). It has recently been shown that the HHV-6A U51-encoded protein is able to bind various CC chemokines in vitro (34). In addition, the RCMV R78 gene was found to play an important role in viral replication in vitro as well as in vivo (2). Like all other sequenced betaherpesviruses, RCMV does not possess ORFs with significant sequence similarity to the US27 and US28 GPCR-like genes of HCMV (12).
(ii) An ORF encoding an MHC class I homolog.
RCMV contains an ORF putatively encoding a homolog of MHC class I heavy chains. This ORF, r144 (4), possesses positional as well as sequence similarity to the MCMV m144 gene (37). We recently reported that an r144-deleted RCMV strain shows similar replication characteristics as wild-type RCMV both in vitro and in immunocompromised rats (4). In contrast, an m144-deleted MCMV strain was shown to be attenuated during the primary phase of infection in mice (18).
(iii) An ORF encoding a CC chemokine homolog.
Previously, genes encoding homologs of chemokines have been identified in both HCMV (UL146, UL147, and UL152) (9) and MCMV (m131/129) (19, 29, 30). The HCMV-encoded chemokine homologs show similarity to CXC (or α-) chemokines, whereas the MCMV m131/129-encoded protein is more related to CC (or β-) chemokines. The MCMV-encoded chemokine homolog was reported to be produced from a transcript in which the m131 ORF is spliced at its 3′ end to the downstream located m129 ORF (19, 29, 30). Remarkably, RCMV possesses an ORF at a position congruent to that of MCMV m131 with limited similarity to both m131 and m129 (Fig. 3). A study of m131-deleted MCMV strains indicated that the m131/129-encoded polypeptide may function as a chemokine agonist by recruiting leukocytes to the sites of infection (19).
FIG. 3.
Alignment of the amino acid sequences predicted to be encoded by RCMV r131 and MCMV m131/129. The alignment was carried out by using a CLUSTAL W Multiple Sequence Alignment Program (version 1.7; Human Genome Sequencing Center, Houston, Tex. [http://dot.imgen.bcm.tmc.edu:9331/multi-align/multi-align.html]) (45). Blocks of identical (white letters in black boxes) and similar (white letters in grey boxes) residues were generated with program BOXSHADE (version 3.21; The EMBnet Foundation, The Netherlands [http://www.ch.embnet.org/software/BOX_form.html]), with the fraction of sequences that must agree for shading set to 1. Numbers to the left of the sequences indicate the positions of amino acid residues within the polypeptides. Cysteine residues that are conserved among CC chemokines are denoted above the sequence by the letter C. The part of the m131/129-derived amino acid sequence that is encoded by either ORF m131 or ORF m129 is also shown. The m131/129-encoded amino acid sequence was taken from MacDonald et al. (29).
RCMV ORF r127 shows similarity to parvovirus rep genes.
ORF r127 is unique among the CMVs: a positional and/or sequence homolog of this ORF was found in neither HCMV nor MCMV. Surprisingly, a TBLASTN database search revealed that the amino acid sequence that was deduced from r127 has similarity to the sequences of NS1 (nonstructural protein 1) or Rep proteins that are encoded by the rep genes of parvoviruses. These viruses have single-stranded DNA genomes with a length of approximately 5 kb. The Parvovirinae subfamily of the parvoviruses consists of three genera, Dependovirus, Parvovirus, and Erythrovirus. The dependoviruses or adeno-associated viruses (AAVs) require helper functions which can be supplied either by genotoxic stimuli or by coinfecting viruses, like adenovirus, HSV-1, HSV-2, CMV, and pseudorabies virus (for a review, see reference 6). These helper functions are needed for productive infection and rescue of viruses that are integrated into the host's genome. Unlike the AAVs, the members of the Parvovirus genus are all capable of autonomous replication and can be pathogenic. The RCMV r127-derived amino acid sequence displays highest similarity with the sequence of the goose parvovirus (GPV) NS1 protein (53). Lower similarities were observed with the corresponding sequences of other parvoviruses, like Barbarie duck parvovirus (53) and AAV-5 (1). Interestingly, the r127-encoded amino acid sequence also showed similarity to the sequence encoded by the U94 gene of HHV-6A (46). A homolog of the U94 gene, which displayed the highest degree of similarity with the rep gene of AAV-2 (41, 46), was also found at a congruent position in the genome of HHV-6B (17, 22). Remarkably, despite the generally close genetic conservation between HHV-6A, HHV-6B, and HHV-7, a U94 homolog was not detected in the genome of HHV-7 (36). ORF 94 is one of only six ORFs (DR3, U6, U9, U22, U83, and U94) that are conserved between HHV-6A and HHV-6B but not HHV-7 (17). It is, therefore, surprising that ORF U94 not only conserves sequence but also genomic position with RCMV r127. In the genomes of HHV-6A and -6B, ORF U94 is located immediately 5′ of the U95 ORF, running from right to left in the direction opposite to that of U95. RCMV r127 is similarly situated with regard to the RCMV homolog of U95, r128. The conserved location as well as orientation of the r127 and U94 genes in their respective genomes indicates that these genes may have diverged from a common ancestral betaherpesvirus genome. A multiple sequence alignment of the amino acid sequences encoded by r127, HHV-6A U94 and GPV rep is shown in Fig. 4. In comparison with the NS1 amino acid sequence, both the r127 and U94 sequences are truncated at their carboxyl termini. The r127-derived sequence is also truncated at its amino terminus compared to the other two sequences. Strongly conserved regions among the three amino acid sequences include sequences that represent putative nucleoside triphosphate binding helicase motifs termed A, B, B', and C (Fig. 4) (21, 26).
FIG. 4.
Alignment of the amino acid sequences predicted to be encoded by RCMV r127, GPV NS1, and HHV-6A U94. The alignment was carried out by using a CLUSTAL W Multiple Sequence Alignment Program (45). Numbers to the left of the sequences indicate the positions of amino acid residues within the polypeptides. Blocks of identical (white letters in black boxes) and similar (white letters in grey boxes) residues were generated with program BOXSHADE (version 3.21), with the fraction of sequences that must agree for shading set to 0.5. The sequence motifs depicted with A, B, B', and C represent strongly conserved putative NTP binding helicase regions (21, 26). The sequences encoded by GPV NS1 and HHV-6A U94 were from Zadori et al. (53) and Thomson et al. (46), respectively.
Rep proteins have been demonstrated to play an essential role in the parvovirus replication cycle, with activities ranging from repression and activation of viral and cellular promoters to site-specific integration into the host genome (6). The HHV-6A U94-encoded protein (RepH6) was found to have a conserved function with respect to its AAV counterpart, since it was shown to complement the replication of Rep-defective AAV-2 mutants (47). In addition, RepH6 was reported to be expressed in the latent phase of HHV-6A infection in vivo, indicating a possible role of this protein in the regulation of latency (38). Whether a similar, important function can be attributed to the RCMV r127 gene product will have to be answered by future investigations.
Nucleotide sequence accession number.
The nucleotide and amino acid sequences discussed in this paper have been deposited in the GenBank database under accession number AF232689.
Acknowledgments
We thank Saskia van der Vlies, Audrey Dasi, Mohamed Siad Farah, and Jasper van Grunsven for technical assistance, Patrick Beisser for helpful discussions, and Suzanne Kaptein and Wil Loenen for critical reading of the manuscript.
REFERENCES
- 1.Bantel-Schaal U, Delius H, Schmidt R, zur Hausen H. Human adeno-associated virus type 5 is only distantly related to other known primate helper-dependent parvoviruses. J Virol. 1999;73:939–947. doi: 10.1128/jvi.73.2.939-947.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Beisser P S, Grauls G, Bruggeman C A, Vink C. Deletion of the R78 G protein-coupled receptor gene from rat cytomegalovirus results in an attenuated, syncytium-inducing mutant strain. J Virol. 1999;73:7218–7230. doi: 10.1128/jvi.73.9.7218-7230.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beisser P S, Kaptein S J F, Beuken E, Bruggeman C A, Vink C. The Maastricht strain and England strain of rat cytomegalovirus represent different betaherpesvirus species rather than strains. Virology. 1998;246:341–351. doi: 10.1006/viro.1998.9196. [DOI] [PubMed] [Google Scholar]
- 4.Beisser P S, Kloover J S, Grauls G E L M, Blok M J, Bruggeman C A, Vink C. The r144 major histocompatibility complex class I-like gene of rat cytomegalovirus is dispensable for both acute and long-term infection in the immunocompromised host. J Virol. 2000;74:1045–1050. doi: 10.1128/jvi.74.2.1045-1050.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beisser P S, Vink C, van Dam J G, Grauls G, Vanherle S J V, Bruggeman C A. The R33 G protein-coupled receptor gene of rat cytomegalovirus plays an essential role in the pathogenesis of viral infection. J Virol. 1998;72:2352–2363. doi: 10.1128/jvi.72.3.2352-2363.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berns K I. Parvoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2173–2197. [Google Scholar]
- 7.Beuken E, Grauls G, Bruggeman C A, Vink C. The rat cytomegalovirus R32 gene encodes a virion-associated protein that elicits a strong humoral immune response in infected rats. J Gen Virol. 1999;80:2719–2728. doi: 10.1099/0022-1317-80-10-2719. [DOI] [PubMed] [Google Scholar]
- 8.Beuken E, Slobbe R, Bruggeman C A, Vink C. Cloning and sequence analysis of the genes encoding DNA polymerase, glycoprotein B, ICP18.5 and major DNA-binding protein of rat cytomegalovirus. J Gen Virol. 1996;77:1559–1562. doi: 10.1099/0022-1317-77-7-1559. [DOI] [PubMed] [Google Scholar]
- 9.Cha T A, Tom E, Kemble G W, Duke G M, Mocarski E S, Spaete R R. Human cytomegalovirus clinical isolates carry at least 19 genes not found in laboratory strains. J Virol. 1995;70:78–83. doi: 10.1128/jvi.70.1.78-83.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang Y, Jeang K, Lietman T, Hayward G S. Structural organization of the spliced immediate-early gene complex that encodes the major acidic nuclear (IE1) and transactivator (IE2) proteins of african green monkey cytomegalovirus. J Biomed Sci. 1995;2:105–130. doi: 10.1007/BF02253062. [DOI] [PubMed] [Google Scholar]
- 11.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison III C A, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 12.Chee M S, Satchwell S C, Preddie E, Weston K M, Barrell B G. Human cytomegalovirus encodes three G protein-coupled receptor homologues. Nature (London) 1990;344:774–777. doi: 10.1038/344774a0. [DOI] [PubMed] [Google Scholar]
- 13.Colberg-Poley A M, Santomenna L D, Harlow P P, Benfield P A, Tenney D J. Human cytomegalovirus US3 and UL36-38 immediate early proteins regulate gene expression. J Virol. 1992;66:95–105. doi: 10.1128/jvi.66.1.95-105.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cranmer L D, Clark C L, Morello C S, Farrell H E, Rawlinson W D, Spector D H. Identification, analysis, and evolutionary relationships of the putative murine cytomegalovirus homologs of the human cytomegalovirus UL82 (pp71) and UL83 (pp65) matrix phosphoproteins. J Virol. 1996;70:7929–7939. doi: 10.1128/jvi.70.11.7929-7939.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crnkovic-Mertens I, Messerle M, Milotic I, Szepan U, Kucic N, Krmpotic A, Jonjic S, Koszinowski U H. Virus attenuation after deletion of the cytomegalovirus Fc receptor gene is not due to antibody control. J Virol. 1998;72:1377–1382. doi: 10.1128/jvi.72.2.1377-1382.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Davis-Poynter N J, Lynch D M, Vally H, Shellam G R, Rawlinson W D, Barrell B G, Farrell H E. Identification and characterization of a G protein-coupled receptor homolog encoded by murine cytomegalovirus. J Virol. 1997;71:1521–1529. doi: 10.1128/jvi.71.2.1521-1529.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dominguez G, Dambaugh T R, Stamey F R, Dewhurst S, Inoue N, Pellett P E. Human herpesvirus 6B genome sequence: coding content and comparison with human herpesvirus 6A. J Virol. 1999;73:8040–8052. doi: 10.1128/jvi.73.10.8040-8052.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farrell H E, Vally H, Lynch D M, Fleming P, Shellam G R, Scalzo A A, Davis-Poynter N J. Inhibition of natural killer cells by a cytomegalovirus MHC class I homologue in vivo. Nature (London) 1997;386:510–514. doi: 10.1038/386510a0. [DOI] [PubMed] [Google Scholar]
- 19.Fleming P, Davis-Poynter N, Degli-Esposti M, Densley E, Papadimitriou J, Shellam G, Farrell H. The murine cytomegalovirus chemokine homolog, m131/129, is a determinant of viral pathogenicity. J Virol. 1999;73:6800–6809. doi: 10.1128/jvi.73.8.6800-6809.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gompels U A, Nicholas J, Lawrence G, Jones M, Thomson B J, Martin M E D, Efstathiou S, Craxton M, Macaulay H A. The DNA sequence of human herpesvirus-6: structure, coding content, and genome evolution. Virology. 1995;209:29–51. doi: 10.1006/viro.1995.1228. [DOI] [PubMed] [Google Scholar]
- 21.Gorbalenya A E, Koonin E V, Wolf Y I. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 1990;262:145–148. doi: 10.1016/0014-5793(90)80175-i. [DOI] [PubMed] [Google Scholar]
- 22.Isegawa Y, Mukai T, Nakano K, Kagawa M, Chen J, Mori Y, Sunagawa T, Kawanishi K, Sashihara J, Hata A, Zou P, Kosuge H, Yamanishi K. Comparison of the complete DNA sequences of human herpesvirus 6 variants A and B. J Virol. 1999;73:8053–8063. doi: 10.1128/jvi.73.10.8053-8063.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Isegawa Y, Ping Z, Nakano K, Sugimoto N, Yamanishi K. Human herpesvirus 6 open reading frame U12 encodes a functional β-chemokine receptor. J Virol. 1998;72:6104–6112. doi: 10.1128/jvi.72.7.6104-6112.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iskenderian A C, Huang L, Reilly A, Stenberg R M, Anders D G. Four of eleven loci required for transient complementation of human cytomegalovirus DNA replication cooperate to activate expression of replication genes. J Virol. 1996;70:383–392. doi: 10.1128/jvi.70.1.383-392.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Keil G M, Ebeling-Keil A, Koszinowsky U. Immediate-early genes of murine cytomegalovirus: location, transcripts, and translation products. J Virol. 1987;61:526–533. doi: 10.1128/jvi.61.2.526-533.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koonin E V. A common set of conserved motifs in a vast variety of putative nucleic acid-dependent ATPases including MCM proteins involved in the initiation of eukaryotic DNA replication. Nucleic Acids Res. 1993;21:2541–2547. doi: 10.1093/nar/21.11.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kouzarides T, Bankier A T, Satchwell S C, Preddie E, Barrell B G. An immediate early gene of human cytomegalovirus encodes a potential membrane glycoprotein. Virology. 1988;165:151–164. doi: 10.1016/0042-6822(88)90668-x. [DOI] [PubMed] [Google Scholar]
- 28.Littler E, Stuart A D, Chee M S. Human cytomegalovirus UL97 open reading frame encodes a protein that phosphorylates the antiviral nucleoside analogue ganciclovir. Nature (London) 1992;358:160–162. doi: 10.1038/358160a0. [DOI] [PubMed] [Google Scholar]
- 29.MacDonald M R, Burney M W, Resnick S B, Virgin H W., IV Spliced mRNA encoding the murine cytomegalovirus chemokine predicts a β chemokine of novel structure. J Virol. 1999;73:3682–3691. doi: 10.1128/jvi.73.5.3682-3691.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.MacDonald M R, Li X-Y, Virgin H W., IV Late expression of a β chemokine homolog by murine cytomegalovirus. J Virol. 1997;71:1671–1678. doi: 10.1128/jvi.71.2.1671-1678.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Margulies B J, Browne H, Gibson W. Identification of the human cytomegalovirus G protein-coupled receptor homologue encoded by UL33 in infected cells and enveloped virus particles. Virology. 1996;225:111–125. doi: 10.1006/viro.1996.0579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meijer H, Dreesen J C, van Boven C P. Molecular cloning and restriction endonuclease mapping of the rat cytomegalovirus genome. J Gen Virol. 1986;67:1327–1342. doi: 10.1099/0022-1317-67-7-1327. [DOI] [PubMed] [Google Scholar]
- 33.Messerle M, Keil G M, Koszinowski U H. Structure and expression of murine cytomegalovirus immediate-early gene 2. J Virol. 1991;65:1638–1643. doi: 10.1128/jvi.65.3.1638-1643.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Milne R S B, Mattick C, Nicholson L, Devaray P, Alcami A, Gompels U. RANTES binding and down-regulation by a novel human herpesvirus-6 β chemokine receptor. J Immunol. 2000;164:2396–2404. doi: 10.4049/jimmunol.164.5.2396. [DOI] [PubMed] [Google Scholar]
- 35.Myers E, Miller W. Optimal alignments in linear space. CABIOS. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- 36.Nicholas J. Determination and analysis of the complete nucleotide sequence of human herpesvirus 7. J Virol. 1996;70:5975–5989. doi: 10.1128/jvi.70.9.5975-5989.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rawlinson W D, Farrell H E, Barrell B G. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70:8833–8849. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rotola A, Ravaioli T, Gonelli A, Dewhurst S, Cassai E, Di Luca D. U94 of human herpesvirus 6 is expressed in latently infected peripheral blood mononuclear cells and blocks viral gene expression in transformed lymphocytes in culture. Proc Natl Acad Sci USA. 1998;95:13911–13916. doi: 10.1073/pnas.95.23.13911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sandford G R, Burns W H. Rat cytomegalovirus has a unique immediate early gene enhancer. Virology. 1996;222:310–317. doi: 10.1006/viro.1996.0428. [DOI] [PubMed] [Google Scholar]
- 40.Sandford G R, Ho K, Burns W H. Characterization of the major locus of immediate early genes of rat cytomegalovirus. J Virol. 1993;67:4093–4103. doi: 10.1128/jvi.67.7.4093-4103.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Srivastava A, Lusby E W, Berns K I. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J Virol. 1983;45:555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stasiak P C, Mocarski E S. Transactivation of the cytomegalovirus ICP36 gene promoter requires the alpha gene product TRS1 in addition to IE1 and IE2. J Virol. 1992;66:1050–1058. doi: 10.1128/jvi.66.2.1050-1058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stenberg R M, Depto A S, Fortney J, Nelson J. Regulated expression of early and late RNAs and proteins from the human cytomegalovirus immediate early gene region. J Virol. 1989;63:2699–2708. doi: 10.1128/jvi.63.6.2699-2708.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thale R, Lucin P, Schneider K, Eggers M, Koszinowski U H. Identification and expression of a murine cytomegalovirus early gene coding for an Fc receptor. J Virol. 1994;68:7757–7765. doi: 10.1128/jvi.68.12.7757-7765.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomson B J, Efstathiou S, Honess R W. Acquisition of the human adeno-associated virus type-2 rep gene by human herpesvirus type-6. Nature (London) 1991;351:78–80. doi: 10.1038/351078a0. [DOI] [PubMed] [Google Scholar]
- 47.Thomson B J, Weindler F W, Gray D, Schwaab V, Heilbronn R. Human herpesvirus 6 (HHV-6) is a helper virus for adeno-associated virus type 2 (AAV-2) and the AAV-2 rep gene homologue in HHV-6 can mediate AAV-2 DNA replication and regulate gene expression. Virology. 1994;204:304–311. doi: 10.1006/viro.1994.1535. [DOI] [PubMed] [Google Scholar]
- 48.Vink C, Beisser P S, Bruggeman C A. Molecular mimicry by cytomegaloviruses. Function of cytomegalovirus-encoded homologues of G protein-coupled receptors, MHC class I heavy chains and chemokines. Intervirology. 1999;42:342–349. doi: 10.1159/000053970. [DOI] [PubMed] [Google Scholar]
- 49.Vink C, Beuken E, Bruggeman C A. Structure of the rat cytomegalovirus genome termini. J Virol. 1996;70:5221–5229. doi: 10.1128/jvi.70.8.5221-5229.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vink C, Beuken E, Bruggeman C A. Cloning and functional characterization of the origin of lytic-phase DNA replication of rat cytomegalovirus. J Gen Virol. 1997;78:2963–2973. doi: 10.1099/0022-1317-78-11-2963. [DOI] [PubMed] [Google Scholar]
- 51.Winkler M, Rice S A, Stamminger T. UL69 of human cytomegalovirus, an open reading frame with homology to ICP27 of herpes simplex virus, encodes a transactivator of gene expression. J Virol. 1994;68:3943–3954. doi: 10.1128/jvi.68.6.3943-3954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu C A, Carlson M E, Henry S C, Shanley J D. The murine cytomegalovirus M25 open reading frame encodes a component of the tegument. Virology. 1999;262:265–276. doi: 10.1006/viro.1999.9942. [DOI] [PubMed] [Google Scholar]
- 53.Zadori Z, Stefancsik R, Rauch T, Kisary J. Analysis of the complete nucleotide sequences of goose and muscovy duck parvoviruses indicates common ancestral origin with adeno-associated virus 2. Virology. 1995;212:562–573. doi: 10.1006/viro.1995.1514. [DOI] [PubMed] [Google Scholar]
- 54.Ziegler H, Thale R, Lucin P, Muranyi W, Flohr T, Hengel H, Farrell H, Rawlinson W, Koszinowski U H. A mouse cytomegalovirus glycoprotein retains MHC class I complexes in the ERGIC/cis-Golgi compartments. Immunity. 1997;6:57–66. doi: 10.1016/s1074-7613(00)80242-3. [DOI] [PubMed] [Google Scholar]