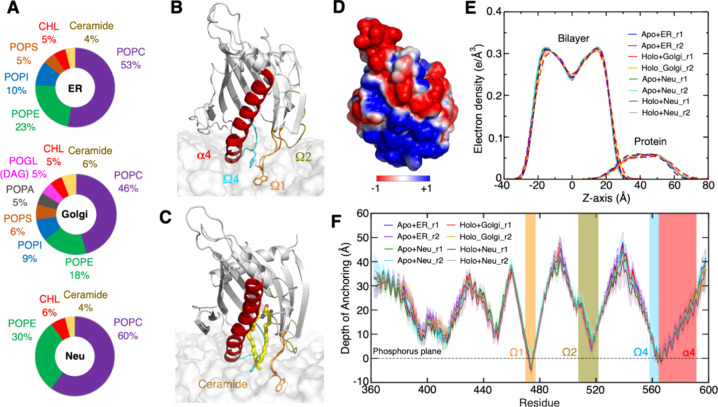
Figure 2.
Binding mode of apo and holo START on ER- and Golgi-like bilayers from molecular dynamics simulation. (A) Lipid compositions in ER-, Golgi-like, and neutral bilayers. (B,C) MD snapshots of the bilayer-bound apo and holo START on ER- and Golgi-like bilayers, respectively. (D) Electrostatic surface potential of apo START mapped on the molecular surface (negative: red, positive: blue) (E) Electron density profiles of the bilayers and protein from MD simulations including both replicas (r1 and r2) in each case, calculated for the last 500 ns of the simulations. (F) Depth of insertion of the Cβ of each START amino acid (Cα of glycines) into the bilayers for all systems and replicas, calculated from the time that the START domain binds to the bilayer to the end of the simulations (see Materials and Methods).