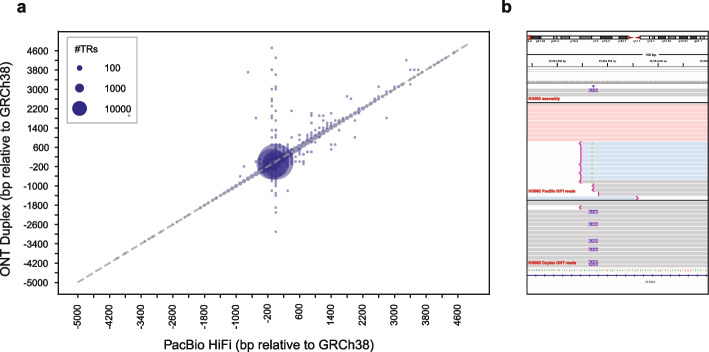
Fig. 2.
a Comparison of LongTR genotypes on ONT Duplex vs. PacBio HiFi data. For each call, we computed the average of the length of each allele relative to the GRCh38 reference. The x-axis gives the calls using PacBio data, and the y-axis gives the calls using ONT Duplex data. Bubble size scales with the number of calls at each coordinate. b IGV [20] screenshot comparing PacBio HiFi reads vs. ONT Duplex reads at a TR genotyped by LongTR. The top window shows the assembly alignment, the middle window shows aligned HiFi reads, and the bottom window shows aligned ONT Duplex reads at a (AGTAAATAATG)n VNTR. All data is aligned to GRCh38. Red and blue denote PacBio HiFi reads from the two haplotypes of HG002 based on haplotag information. Gray reads have no haplotag information. HiFi reads were clipped at the large repeat insertion, resulting in an incorrect genotype call