Abstract
Perry syndrome (PS) is a rare autosomal dominant disease characterized by parkinsonism, central hypoventilation, weight loss and depression and is caused by pathogenic mutations in the dynactin subunit 1 (DCTN1) gene (encoding p150glued protein). To date, only two cases have been reported in Latin America, specifically in Colombia and Argentina. The present study, to the best of our knowledge, reports the first recorded Mexican family with PS. The clinical features of the proband and a family history of early parkinsonism led to the suspicion of PS. The pathogenic variant NM_004082:c.212G>A, causing a (p.Gly71Glu) mutation in the p150glued protein, was identified in exon 2 of the DCTN1 gene by exome sequencing, confirming the diagnosis of PS. (p.Gly71Glu) has been previously identified in at least 4 cases of PS from different ethnic backgrounds. Genetic counseling was provided to the available family members. To clarify the impact of the (p.Gly71Glu) variant on the structure and function of the cytoskeleton-associated protein Gly rich (CAP-Gly) domain of p150glued, Glu71 mutated CAP-Gly domains were modeled and compared with the wild-type. It was hypothesized that the larger and more charged side chain of Glu may induce conformational and electrostatic changes, imposing a conformational restriction on the peptide backbone that would affect interaction with the p150glued protein partners, causing dysfunction in the dynactin protein complex.
Keywords: Perry syndrome, DCTN1, pathogenic variant, parkinsonism, in silico analysis
Introduction
Perry syndrome (PS; Mendelian inheritance in man ID, 168605) is a rare hereditary progressive neurodegenerative disease first described in 1975 by Perry et al (1) in Canada. PS is characterized by early onset parkinsonism, central hypoventilation, severe weight loss and neuropsychiatric features (mainly apathy and depression), which begin at ~48 years of age. Psychiatric alterations and parkinsonism are typically the first symptoms to appear, followed by weight loss (2). Post-mortem brain examination of PS cases revealed a severe degeneration in the substantia nigra with the presence of TAR DNA binding protein (TDP)-43 inclusions in the basal ganglia and brain stem (3). However, the observed inclusion pattern was different from those reported in other neurological diseases involving TDP-43 proteinopathy (4).
PS is caused by pathogenic variants in the dynactin subunit 1 (DCTN1) gene, located at locus 2p13.1 (5,6). The DCTN1 gene was characterized in 1994(7) and encodes the p150glued protein, the largest subunit of the macromolecular complex, dynactin. Dynactin has 10 subunits and is involved in a number of diverse cellular functions, including endoplasmic reticulum-to-Golgi transport, centripetal movement of lysosomes and endosomes, spindle formation, chromosome movement, nuclear positioning and axonogenesis (7,8). PS is inherited as an autosomal dominant trait (5,9), with an established age-dependent penetrance and a certain variability in clinical features (10). Most pathogenic variants associated with PS are located on exon 2 of the gene encoding the cytoskeleton-associated protein Gly rich (CAP-Gly) domain of p150glued. Pathogenic variants of DCTN1 also cause distal hereditary motor neuronopathy type 7B (also known as distal spinal and bulbar muscular atrophy) (11), progressive supranuclear palsy (PSP)-like phenotype frontotemporal dementia (12) and motor neuron disease/amyotrophic lateral sclerosis. Certain individuals also present with overlapping phenotypes (10).
PS has no cure and requires multidisciplinary clinical management, depending on the clinical features of each case. For instance, parkinsonism is treated with dopaminergic therapy, psychiatric manifestations are treated with antidepressants and weight loss is treated with hypercaloric and hyperproteic diets. In addition, ventilatory support (invasive or non-invasive) improves the survival rate of patients with central hypoventilation (9,10). However, affected individuals typically die from respiratory complications and the average survival rate is 5 years following diagnosis (10).
The present study described the first (to the best of our knowledge) Mexican family diagnosed with PS. Molecular confirmation of this diagnosis was obtained using a mini-exome sequencing panel. The potential impact of the identified DCTN1 mutation on the structure and function of the p150glued protein was assessed using in silico analysis.
Patient and methods
Case report
Clinical evaluation was performed in August 2016 at the Department of Medical Genetics of the National Medical Center, ‘La Raza’ (Mexico City, Mexico), which is part of the Mexican Social Security Institute. The proband was a Mexican mestizo male aged 55 years at the time of consultation. The proband's family was originally from the state of Hidalgo, Mexico. The proband's main symptoms were chronic fatigue for 6 years and weight loss in the previous year at a rate of 1 kg per month, the body mass index at presentation was 14.66 kg/m2 (normal range, 18.5-24.9 kg/m2), which is considered severely underweight. The proband also reported difficulty performing daily activities due to bradykinesia, gait disturbances, dyskinesias and tremors. Physical examination revealed that the proband looked older than their chronological age with several facial wrinkles and gray hair. The proband also had a steppage gait, marked reduction in subcutaneous fat, expressionless facies and scanning speech. Facial hypertrichosis, horizontal saccadic eye movements, slight blinking and stiffness in the upper and lower extremities were also observed. The proband showed no clinical signs of depression and did not report any suicide attempts.
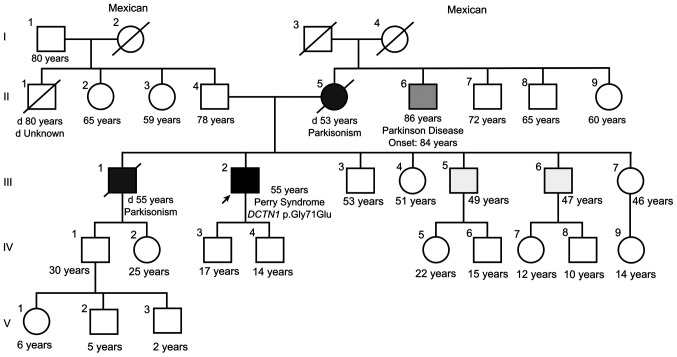
The family history (Fig. 1) indicated that the proband's mother (II.5) and one of the proband's brothers (III.1) had died from complications of Parkinson's disease (PD) at the ages of 53 and 55 years, respectively. A maternal uncle had also developed late-onset PD (II.6). The proband had two sons (IV.3 and IV.4), aged 17 and 14 years at the time of the present study. The clinical data and family history of the proband led to the suspicion of PS. In addition, a polysomnography study corroborated the presence of central alveolar hypoventilation and alterations in sleep patterns.
Figure 1.
Pedigree of the Mexican family with Perry syndrome. Squares represent male individuals and circles represent females. The arrow indicates the proband case. Black symbols indicate affected individuals. Diagonal lines indicate deceased individuals and the age of death is stated if known.
Mini-exome sequencing
Following genetic counseling, the proband signed an informed consent form for molecular analysis. The analysis was conducted at the Laboratory of Genomic Diagnosis at the National Institute of Genomic Medicine (Mexico City, Mexico). For this, genomic DNA was obtained from EDTA-peripheral blood. DNA was extracted using an AS1010 cartridge and the Maxwell 16 System (Promega Corp.) following the manufacturer's instructions. The quality and integrity of the DNA were evaluated using a NanoDrop 1000 spectrophotometer (Thermo Fisher Scientific, Inc.) and Qubit fluorometer (Thermo Fisher Scientific, Inc.). Exome sequencing was performed using the TruSight One Sequencing Panel kit (Illumina, Inc.) following the manufacturer's instructions. This kit allowed for simultaneous sequencing of exons and splicing regions of 4,813 genes, including the DCTN1 gene and other genes associated with autosomal dominant PD. Sequencing was performed using the MiSeq Instrument and v3 reagents (Illumina, Inc.). A paired-end read of 150 bp and a library pool concentration of 12.5 pM quantified by a Qubit fluorometer (Thermo Fisher Scientific, Inc.) were used. A 100x mean coverage of the targeted regions was obtained. Data analyses were performed using the Trimmomatic tool (13), Burrows-Wheeler Aligner-Maximal Exact Matches (14) and the Genome Analysis Toolkit algorithm (15).
Prediction of the functional impact of the DCTN1 variant
The Sorting Intolerant From Tolerant (SIFT) (16), Polymorphism Phenotyping v2 (PolyPhen-2) (17) and MutationTaster (18) algorithms were used to predict the functional effects of the identified DCTN1 variant. Population databases such as the Genome Aggregation Database (gnomAD; https://gnomad.broadinstitute.org/), 1000 Genomes Project (https://www.internationalgenome.org/) and ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and the literature in PubMed (https://pubmed.ncbi.nlm.nih.gov/) were also reviewed (the search terms used in the databases are provided in the supplementary material).
Computational modeling of the variant
In total, four different models of p150glued bearing the mutation were generated using SWISS-MODEL (19). Models 1, 2, 3 and 4 were generated using the Protein Data Bank (PDB) structures: Solution structure of the CAP-Gly domain in human dynactin 1 (2COY), crystal structure of the C-terminal domain of human EB1 in complex with the CAP-Gly domain of human dynactin-1 (p150-Glued) (2HKQ), three-dimensional structure of cap-gly domain assembled on microtubules determined by magic angle spinning (MAS) NMR spectroscopy (2MPX) and crystal structure of the zink-knuckle 2 domain of human CLIP-170 in complex with p150-Glued (3E2U) as templates. The target sequence bearing the (p.Gly71Glu) variant was defined in each case by aligning the reference sequence of the human p150glued protein (accession no. NP_004073.2) with that of the template molecule. The models were visualized and analyzed using PyMOL software version 1.8 (Schrödinger, LLC) (20). The Voronoia 1.0 program suite was used to detect the internal cavities of the models (21).
Protein electrostatic potential
Using the default settings, the electrostatic potential maps of the mutant model structures and the structures used as templates were constructed using Swiss-PdbViewer 4.1.0(22).
Results
Mini-exome sequencing analysis
Following exome sequencing, the NM_004082.4:c.212G>A variant was identified in exon 2 of DCTN1, causing a Gly substitution for Glu at position 71 of the protein, NP_004073.2:(p.Gly71Glu). Gly71 belongs to the Gly-Lys-Asn-Asp-Gly (GKNDG) motif, which is highly conserved in numerous CAP-Gly domains (23). The Gly71Glu substitution was predicted to be deleterious by the SIFT algorithm (score 0), probably damaging by the PolyPhen-2 algorithm (HumVar score, 0.9) and a cause of disease by the MutationTaster (score, 0.999999997834239) algorithm. This variant has been submitted to ClinVar three times (ClinVar ID: 34242) but was absent from the exome and genome sequencing data submitted to gnomAD (24) and from the Mexican database available through the Franklin by Genoox platform (https://franklin.genoox.com/clinical-db/home). No pathogenic or probable pathogenic variants were identified in genes associated with autosomal dominant PD or other neurological diseases with similar clinical manifestations.
None of the other family members of the proband accepted medical examination or molecular analysis. Since the proband's sons were minors at the time of the study, molecular testing was not offered as per the recommendations of the National Society of Genetic Counselors, the American Academy of Pediatrics, the American College of Medical Genetics and Genomics (ACMG) and the American Society of Human Genetics, which indicate that a pre-symptomatic diagnosis should not be performed on minors (25). At present, both sons remain asymptomatic (last contact March, 2024).
Modeling and in silico evaluation of the Gly71Glu variant
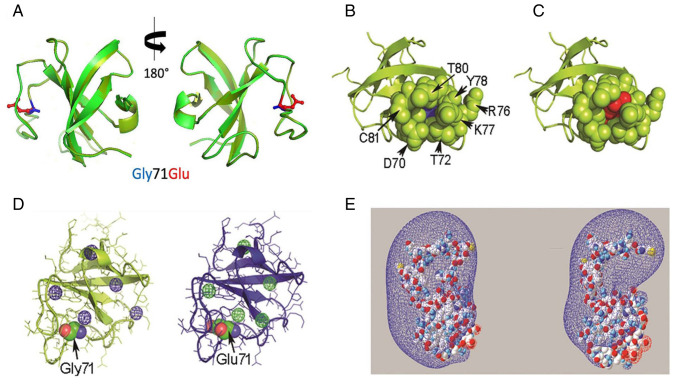
Based on the different reported structures of p150glued, four models of p150glued Gly71Glu were obtained (Figs. 2A and S1). Fig. 2A shows the structural alignment of the wild-type CAP-Gly domain (PDB: 2COY) and the Gly71Glu CAP-Gly domain in Model 1. The spatial orientation and atomic contacts of Glu71 in Model 1 point toward the solvent and occupy a cavity delimited by the side chains of Asp70, Thr72, Lys77, Tyr78, Phe79 and Thr80, as well as the peptide backbone atoms of Tyr78 (Fig. 2A-C). This cavity was empty in the wild-type protein (Fig. 2B). In the structural context of Model 1, Glu71 establishes a pair of H-bonds between its γ carboxylate group, the hydroxyl group of Thr80 and the peptide backbone amide of residue Phe79. A comparison of Models 2 and 4 is provided in Fig. S1. Computational analysis showed that Model 1 had five internal cavities, which was similar to the template (Fig. 2D). A comparison of the cavities in Models 2 and 4 is provided in Fig. S2.
Figure 2.
In silico evaluation of the Gly71Glu variant-spatial relationship of the residue at position 71 of the CAP-Gly domain. (A) Structural alignment of the PDB 2COY template (light green) and Model 1 (dark green). The side chains of Gly71 and Glu71 are displayed in blue and red, respectively. (B) Representative image of residues located spatially close (4 Å) to Gly71 in the wild-type CAP-Gly domain in the free state (PDB 2COY). (C) Equivalent representative image of Glu71 in Model 1. Gly71 and Glu71 are represented as ‘spheres’ in blue and red, respectively. The residues in the proximity of position 71 are shown as green spheres. (D) Different views of the cavities found in PDB 2COY and Model 1 using the Voronoia program suite. The cavities are represented as spheres in a mesh format, in blue or green. (E) Different perspectives of the electrostatic potential map of the PDB 2COY and Model 1 protein structures constructed using Swiss-PdbViewer 4-1-0. The conventional code represents the charge distribution, where blue and red represent the electron-poor and electron-rich regions, respectively. The molecules are represented in a space-filled format, using the CPK color code: Carbon is gray, oxygen is red, nitrogen is blue, sulfur is yellow and hydrogen is white. PDB, protein data bank; CAP-Gly, cytoskeleton-associated protein glycine-rich domain; CPK, Corey-Pauling-Koltun.
Protein electrostatic potential
The electrostatic potential map of the CAP-Gly domain structure in Model 1 is shown in Fig. 2E. The Glu71 variant expanded the electron-rich region in the modeled structure compared with the template. The electrostatic potential maps for Models 2 and 4 are shown in Fig. S3.
Discussion
PS is a rare disease, to the best of our knowledge, the proband reported in this study was the first to be identified in Mexico and the third in Latin America. A summary of the reported PS cases and the identified mutation is shown in Table SI. The first identified case in Latin America harbored the (p.Gly71Arg) pathogenic variant and was reported in Colombia in 2014 by Pretelt et al (26). In 2022, the (p.Gly67Asp) variant was identified in an Argentine family by Silva et al (27). The other confirmed PS cases were mainly of European or Asian origin (28).
The molecular testing using exome sequencing identified the NM_004082:c.212G>A (p.Gly71Glu) pathogenic variant in the DCTN1 gene, confirming the diagnosis of PS in the proband. The (p.Gly71Glu) variant was first identified by Farrer et al (5) in 2009 in a French family with PS. There were no data suggesting French ancestry in the present case; therefore, the variant likely resulted from a de novo mutation in this family.
The proband reported in the present study had three of the four cardinal criteria proposed by Wider and Wszolek (9): Autosomal dominant parkinsonism, weight loss and central hypoventilation. No depression, apathy, social withdrawal or suicidal attempts were reported at the first medical appointment or during the following 2 years. However, a formal psychiatric evaluation of the proband was never performed; therefore, the absence of psychiatric symptoms must be considered with caution. Table SII lists the clinical features of other PS patients with the Gly71Glu variant, as documented in the literature. The clinical features are similar among patients with the same variant. Considering the proband's chronic fatigue in the 6 years prior to diagnosis, the onset of symptoms was likely in the fifth decade of life (~49 years of age), similar to other reported patients harboring the same mutation.
Before the identification of the DCTN1 gene and the possibility of molecular testing, diagnostic confirmation of PS was impossible. The syndrome was classified as Parkinsonism-plus due to alterations in extraocular movements and the presence of pyramidal dysfunction and respiratory disorders (29). The availability of molecular testing using multigene next-generation sequencing-based panels contributes to an earlier diagnosis, confirming the clinical suspicion, identifying the precise molecular defect and ruling out other autosomal dominant parkinsonism diseases (such as variants in MAPT). The identification of the molecular defect allows for more accurate genetic counseling and identification of pre-symptomatic carriers among at-risk asymptomatic relatives (12). Early disease diagnosis may also improve the quality of life of the patient by reducing complications and allowing for timely interventions, such as using non-invasive mechanical ventilation or inserting a diaphragmatic pacemaker, avoiding unnecessary medical and diagnosis procedures for other affected family members.
However, the early detection of PS is impaired by several factors: PS is a rare disease and certain clinical and pathological features overlap with other more common and well-known sporadic neurodegenerative disorders, limiting the rise of clinical suspicion. In these instances, patients would be diagnosed with autosomal dominant parkinsonism or PSP until a marked weight loss is observed, central hypoventilation develops or respiratory failure and death occur (2). For example, two relatives of the proband presented with early parkinsonism (individual II.5, deceased at 53 years of age; and individual III.1, deceased at 55 years of age) and both had been diagnosed with PD. No additional studies were performed to establish the cause of the familial PD and the possibility of PS was not suggested to the family by the attending physician at the time. Additionally, PS may be accompanied by phenotypic heterogeneity among the affected individuals in the same family (30). Caroppo et al (12) reported a family with the (p.Gly71Glu) mutation, in which the affected individuals presented with PS or PSP-like or frontotemporal dementia.
Pathogenic variants of DCTN1 are also associated with different clinical phenotypes. Most variants associated with PS affect the GKNDG motif of the CAP-Gly domain (31), particularly residues 67, 71, 72, 74 and 78. Recently, two new reports of genetic variants outside the CAP-Gly domain in exon 2 were reported. The first case was identified in a female patient without a family history of PS. The variant was demonstrated to be pathogenic based on the clinical features and autopsy findings (32). The second was a 65-year-old female patient who presented with difficulties in falling asleep and personality changes that progressed over four years. Exome sequencing revealed a novel splicing site mutation, c.279+1G>T, in the DCTN1 gene. The variant was classified as pathogenic according to the ACMG Standards and Guidelines (33).
The missense variant (p.Phe52Leu) is associated with late-onset PD, hypoventilation and frontotemporal atrophy (34). By contrast, a variant affecting Gly59 has been associated with distal hereditary motor neuronopathy type VII (11), whereas variants Ile935Met, Ile196Val and Ser111Cys have been associated with susceptibility to amyotrophic lateral sclerosis (35). Certain variants associated with PSP-like syndrome are the same as those identified in PS (5); however, other alleles have also been associated with this phenotype (36).
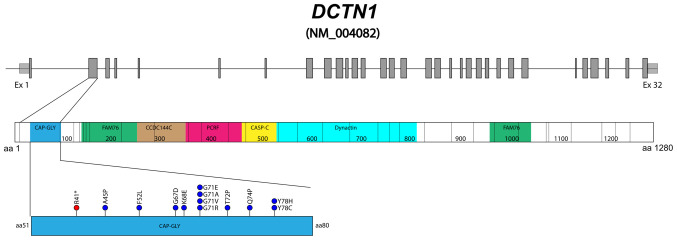
Gly71 appears to be a recurrent site of mutation in DCTN1, since four out of the 14 PS causal variants identified thus far affect residue 71 (p.Gly71Arg/Glu/Val/Ala) (Fig. 3), according to the Human Gene Mutation Database (as of March 2024). The (p.Gly71Glu) variant has been identified in at least three additional patients with PS from different ethnic backgrounds, confirming its pathogenicity (37). In the present study, to clarify the impact of (p.Gly71Glu) on the structure and function of the CAP-Gly domain, the Glu71 mutated CAP-Gly domain was modeled and compared with the wild-type. The model indicated that the presence of Glu induced local changes in the conformation and environment of the CAP-Gly domain. The larger and more charged side chain of Glu resulted in positive volume variation and a negative charge gain at physiological pH at the mutated site. This change imposed a conformational restriction on the peptide backbone, affecting its ability to rearrange appropriately when the CAP-Gly domain interacts with its functional partners (38). It has been shown that the conformational flexibility of the CAP-Gly domain is crucial for the efficient function of p150glued (39). Several regions of the CAP-Gly domain participate in the binding of the p150glued protein partners (26). Among these regions, the conserved GKNDG motif interacts with the acidic-aromatic sequence motifs located at the C-terminus of the protein partners (5).
Figure 3.
Diagram of the DCTN1 gene and p150glued protein. Variants causing Perry syndrome identified in the cytoskeleton-associated protein Gly rich domain (residues 51 to 80, in blue). DCTN1 consists of 32 exons (NM_ 004082) (in gray) and the encoded protein is composed of 1,276 residues. CAP-Gly, cytoskeleton-associated protein glycine-rich domain; FAM76, FAM76 protein domain; CCDC144C, protein coiled-coil region domain; PCRFF, PCRF domain; CASP_C, CASP C terminal domain; Dynactin, Dynein-associated protein domain; DCTN1, dynactin subunit 1.
In the present study, in Models 2 and 4, Gly71 was oriented towards the core of the domain, contacting Phe79, which connected Gly71 with the conserved cluster of aromatic residues (Tyr46, Phe52, Trp57, Tyr78, Phe79 and Phe88), stabilizing the native folding of the CAP-Gly domain. These contacts with Glu71 resulted in the rearrangement of Phe79 in Model 2, which would destabilize the domain by perturbing the interaction network of the conserved aromatic residues present in the folding core. In Model 3, substitution with Glu71 led to less compact folding, characterized by several empty cavities. It has been shown that cavities resulting from packing defects typically destabilize proteins (40) and promote their aggregation (41). As a protein becomes thermodynamically less stable, the probability of adopting non-native conformations prone to aggregation increases. TDP-43 proteinopathy and dynactin aggregates have been detected in all of the examined PS post-mortem brains reported (3-5). In addition, in cells coexpressing the (p.Gly71Ala) mutant and TDP-43, cytoplasmic mislocalization of TDP-43 and aggregates of variable sizes were detected in the cytoplasm and neurites, indicating that mutated DCTN1 induced mislocalization and aggregation of wild-type TDP-43 protein in human neurons (42). In the present study, the constructed electrostatic potential map indicated that Glu71 modified the electrostatic potential of the CAP-Gly domain. However, the effect on charge distribution differed from one model to another. Since the electrostatic potential is a factor that regulates protein-protein interactions (43), the overall change in charge distribution induced by Glu71 would also contribute to disrupting the binding of the CAP-Gly domain to the acidic-aromatic sequence motifs of binding partners (23). In summary, the contribution of the (p.Gly71Glu) variant to the pathogenesis of PS is most likely related to the processes that support the function of proteins, such as folding efficiency, folding stability, protein dynamics, conformational fitting ability and the affinity of interactions with the binding partners (44).
The present study reported the first case of PS identified in Mexico; however, certain limitations hindered a more comprehensive disease description in the proband and their family: Brain images were not available for any of the affected family members and post-mortem brain evaluation was unfeasible. Variant segregation in healthy and symptomatic family members was not possible at the time of the study; however, individual III.5 (living outside of Mexico) was also recently diagnosed and molecularly confirmed with PS (personal communication with a family member).
In conclusion, the present study revealed for the first time that PS is present in the Mexican population, expanding the geographical and ethnic prevalence of this disease and enriching the global dataset of Perry syndrome cases. The clinical features of the proband and the family history of early parkinsonism suggested a diagnosis of PS, which was confirmed by the identification of the pathogenic DCTN1 variant. Unlike the PS cases previously reported, the proband of the present study lacks psychiatric manifestations such as depression and apathy at diagnosis and during the clinical follow-up period. In addition, the in silico modeling of the (p.Gly71Glu) variant allowed us to provide new insight into how this change influences the structural and functional integrity of the cytoskeleton-associated protein complex. We hope this work also raises awareness of rare neurological disorders that are often overlooked in low- and medium-income populations due to the limited availability of genetic testing.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
Funding: No funding was received.
Availability of data and materials
The data generated in the present study are not publicly available due to restrictions in the informed consent form, which does not allow for the unrestricted disclosure of sequencing data, but may be requested from the corresponding author. The information related to the identified DCTN1 variant has been submitted to the ClinVar database (variation ID, 21390; submission ID, 14421829).
Authors' contributions
LFL and ERM performed the clinical evaluations. KCS, CMG and MJO performed the sequencing. CAV, LFL and JGS interpreted the sequencing data and wrote the manuscript. LDPY, URC and GAH analyzed the protein structure and wrote the manuscript. All authors read and approved the final version of the manuscript. CAV, LFL and JGS confirm the authenticity of all the raw data.
Ethics approval and consent to participate
The present study was reviewed and approved by the Ethical Committee of the National Institute of Genomic Medicine in Mexico City, Mexico (approval no. CEI 2018/37) and conducted in accordance with the Code of Ethics of the World Medical Association (Declaration of Helsinki). The proband provided informed consent for molecular testing.
Patient consent for publication
The proband signed an informed consent form for publication of this case.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Perry TL, Bratty PJ, Hansen S, Kennedy J, Urquhart N, Dolman CL. Hereditary mental depression and Parkinsonism with taurine deficiency. Arch Neurol. 1975;32:108–113. doi: 10.1001/archneur.1975.00490440058009. [DOI] [PubMed] [Google Scholar]
- 2.Tsuboi Y, Mishima T, Fujioka S. Perry disease: Concept of a new disease and clinical diagnostic criteria. J Mov Disord. 2021;14:1–9. doi: 10.14802/jmd.20060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wider C, Dickson DW, Stoessl AJ, Tsuboi Y, Chapon F, Gutmann L, Lechevalier B, Calne DB, Personett DA, Hulihan M, et al. Pallidonigral TDP-43 pathology in Perry syndrome. Parkinsonism Relat Disord. 2009;15:281–286. doi: 10.1016/j.parkreldis.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mishima T, Koga S, Lin WL, Kasanuki K, Castanedes-Casey M, Wszolek ZK, Oh SJ, Tsuboi Y, Dickson D. Perry Syndrome: A Distinctive Type of TDP-43 Proteinopathy. J Neuropathol Exp Neurol. 2017;76:676–682. doi: 10.1093/jnen/nlx049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Farrer MJ, Hulihan MM, Kachergus JM, Dächsel JC, Stoessl AJ, Grantier LL, Calne S, Calne DB, Lechevalier B, Chapon F, et al. DCTN1 mutations in Perry syndrome. Nat Genet. 2009;41:163–165. doi: 10.1038/ng.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Holzbaur EL, Tokito MK. Localization of the DCTN1 gene encoding p150Glued to human chromosome 2p13 by fluorescence in situ hybridization. Genomics. 1996;31:398–399. doi: 10.1006/geno.1996.0068. [DOI] [PubMed] [Google Scholar]
- 7.Holzbaur EL, Vallee RB. DYNEINS: Molecular structure and cellular function. Annu Rev Cell Biol. 1994;10:339–372. doi: 10.1146/annurev.cb.10.110194.002011. [DOI] [PubMed] [Google Scholar]
- 8.Gauthier LR, Charrin BC, Borrell-Pagès M, Dompierre JP, Rangone H, Cordelières FP, De Mey J, MacDonald ME, Lessmann V, Humbert S, Saudou F. Huntingtin controls neurotrophic support and survival of neurons by enhancing BDNF vesicular transport along microtubules. Cell. 2004;118:127–138. doi: 10.1016/j.cell.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 9.Wider C, Wszolek ZK. Rapidly progressive familial parkinsonism with central hypoventilation, depression and weight loss (Perry syndrome)-a literature review. Parkinsonism Relat Disord. 2008;14:1–7. doi: 10.1016/j.parkreldis.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 10.Konno T, Ross OA, Teive HAG, Sławek J, Dickson DW, Wszolek ZK. DCTN1-related neurodegeneration: Perry syndrome and beyond. Parkinsonism Relat Disord. 2017;41:14–24. doi: 10.1016/j.parkreldis.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Puls I, Oh SJ, Sumner CJ, Wallace KE, Floeter MK, Mann EA, Kennedy WR, Wendelschafer-Crabb G, Vortmeyer A, Powers R, et al. Distal spinal and bulbar muscular atrophy caused by dynactin mutation. Ann Neurol. 2005;57:687–694. doi: 10.1002/ana.20468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caroppo P, Le Ber I, Clot F, Rivaud-Péchoux S, Camuzat A, De Septenville A, Boutoleau-Bretonnière C, Mourlon V, Sauvée M, Lebouvier T, et al. DCTN1 mutation analysis in families with progressive supranuclear palsy-like phenotypes. JAMA Neurol. 2014;71:208–215. doi: 10.1001/jamaneurol.2013.5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The genome analysis toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1781. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 17.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–289. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–362. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 19.Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rigsby RE, Parker AB. Using the PyMOL application to reinforce visual understanding of protein structure. Biochem Mol Biol Educ. 2016;44:433–437. doi: 10.1002/bmb.20966. [DOI] [PubMed] [Google Scholar]
- 21.Rother K, Hildebrand PW, Goede A, Gruening B, Preissner R. Voronoia: Analyzing packing in protein structures. Nucleic Acids Res. 2009;37(Database issue):D393–D395. doi: 10.1093/nar/gkn769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 23.Weisbrich A, Honnappa S, Jaussi R, Okhrimenko O, Frey D, Jelesarov I, Akhmanova A, Steinmetz MO. Structure-function relationship of CAP-Gly domains. Nat Struct Mol Biol. 2007;14:959–967. doi: 10.1038/nsmb1291. [DOI] [PubMed] [Google Scholar]
- 24.Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature. 2020;581:434–443. doi: 10.1038/s41586-020-2308-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pretelt F, Castañeda Cardona C, Tacik P, Ross OA, Wszolek ZK. Latin America's first case of Perry syndrome and a new treatment option for respiratory insufficiency. J Neurol. 2014;261:620–621. doi: 10.1007/s00415-014-7262-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silva E, Itzcovich T, Niikado M, Caride A, Fernández E, Vázquez JC, Romorini L, Marazita M, Sevlever G, Martinetto H, Surace EI. Perry disease in an Argentine family due to the DCTN1 p.G67D variant. Parkinsonism Relat Disord. 2022;97:63–64. doi: 10.1016/j.parkreldis.2022.03.009. [DOI] [PubMed] [Google Scholar]
- 28.Tacik P, Fiesel FC, Fujioka S, Ross OA, Pretelt F, Castañeda Cardona C, Kidd A, Hlavac M, Raizis A, Okun MS, et al. Three families with Perry syndrome from distinct parts of the world. Parkinsonism Relat Disord. 2014;20:884–888. doi: 10.1016/j.parkreldis.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Denson MA, Wszolek ZK. Familial parkinsonism: Our experience and review. Parkinsonism Relat Disord. 1995;1:35–46. doi: 10.1016/1353-8020(95)00010-4. [DOI] [PubMed] [Google Scholar]
- 30.Dulski J, Koga S, Prudencio M, Tipton PW, Ali S, Strongosky AJ, Rose JH, Parrales ZA, Dunmore JA, Jansen-West K, et al. Perry syndrome: Novel DCTN1 mutation in a large kindred and first observation of prodromal disease. Parkinsonism Relat Disord. 2023;112(105481) doi: 10.1016/j.parkreldis.2023.105481. [DOI] [PubMed] [Google Scholar]
- 31.Mishima T, Ishikawa T, Imamura K, Kondo T, Koshiba Y, Takahashi R, Takahashi J, Watanabe A, Fujii N, Tsuboi Y, Inoue H. Cytoplasmic aggregates of dynactin in iPSC-derived tyrosine hydroxylase-positive neurons from a patient with Perry syndrome. Parkinsonism Relat Disord. 2016;30:67–72. doi: 10.1016/j.parkreldis.2016.06.007. [DOI] [PubMed] [Google Scholar]
- 32.Dulski J, Koga S, Liberski PP, Sitek EJ, Butala AA, Sławek J, Dickson DW, Wszolek ZK. Perry disease: Expanding the genetic basis. Mov Disord Clin Pract. 2023;10:1136–1142. doi: 10.1002/mdc3.13764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tian W, Yao L, Shi G, Dai R, Cao L. A novel DCTN1 mutation causing perry syndrome leads to abnormal splicing of mRNA: Genetic and functional analyses. Acta Neurol Belg. 2024;124:661–663. doi: 10.1007/s13760-023-02368-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Araki E, Tsuboi Y, Daechsel J, Milnerwood A, Vilarino-Guell C, Fujii N, Mishima T, Oka T, Hara H, Fukae J, Farrer MJ. A novel DCTN1 mutation with late-onset parkinsonism and frontotemporal atrophy. Mov Disord. 2014;29:1201–1204. doi: 10.1002/mds.25833. [DOI] [PubMed] [Google Scholar]
- 35.Cady J, Allred P, Bali T, Pestronk A, Goate A, Miller TM, Mitra RD, Ravits J, Harms MB, Baloh RH. Amyotrophic lateral sclerosis onset is influenced by the burden of rare variants in known amyotrophic lateral sclerosis genes. Ann Neurol. 2015;77:100–113. doi: 10.1002/ana.24306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gustavsson EK, Trinh J, Guella I, Szu-Tu C, Khinda J, Lin CH, Wu RM, Stoessl J, Appel-Cresswell S, McKeown M, et al. DCTN1 p.K56R in progressive supranuclear palsy. Parkinsonism Relat Disord. 2016;28:56–61. doi: 10.1016/j.parkreldis.2016.04.025. [DOI] [PubMed] [Google Scholar]
- 37.Dulski J, Cerquera-Cleves C, Milanowski L, Kidd A, Sitek EJ, Strongosky A, Vanegas Monroy AM, Dickson DW, Ross OA, Pentela-Nowicka J, et al. Clinical, pathological and genetic characteristics of Perry disease-new cases and literature review. Eur J Neurol. 2021;28:4010–4021. doi: 10.1111/ene.15048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayashi I, Wilde A, Mal TK, Ikura M. Structural basis for the activation of microtubule assembly by the EB1 and p150Glued complex. Mol Cell. 2005;19:449–460. doi: 10.1016/j.molcel.2005.06.034. [DOI] [PubMed] [Google Scholar]
- 39.Yan S, Zhang H, Hou G, Ahmed S, Williams JC, Polenova T. Internal dynamics of dynactin CAP-Gly is regulated by microtubules and plus end tracking protein EB1. J Biol Chem. 2015;290:1607–1622. doi: 10.1074/jbc.M114.603118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zwarycz AS, Fossat M, Akanyeti O, Lin Z, Rosenman DJ, Garcia AE, Royer CA, Mills KV, Wang C. V67L mutation fills an internal cavity to stabilize RecA Mtu Intein. Biochemistry. 2017;56:2715–2722. doi: 10.1021/acs.biochem.6b01264. [DOI] [PubMed] [Google Scholar]
- 41.Fernández A, Berry RS. Proteins with H-bond packing defects are highly interactive with lipid bilayers: Implications for amyloidogenesis. Proc Natl Acad Sci USA. 2003;100:2391–2396. doi: 10.1073/pnas.0335642100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Deshimaru M, Kinoshita-Kawada M, Kubota K, Watanabe T, Tanaka Y, Hirano S, Ishidate F, Hiramoto M, Ishikawa M, Uehara Y, et al. DCTN1 Binds to TDP-43 and Regulates TDP-43 Aggregation. Int J Mol Sci. 2021;22(3985) doi: 10.3390/ijms22083985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Z, Witham S, Alexov E. On the role of electrostatics in protein-protein interactions. Phys Biol. 2011;8(035001) doi: 10.1088/1478-3975/8/3/035001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vendruscolo M, Zurdo J, MacPhee CE, Dobson CM. Protein folding and misfolding: A paradigm of self-assembly and regulation in complex biological systems. Philos Trans A Math Phys Eng Sci. 2003;361:1205–1222. doi: 10.1098/rsta.2003.1194. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data generated in the present study are not publicly available due to restrictions in the informed consent form, which does not allow for the unrestricted disclosure of sequencing data, but may be requested from the corresponding author. The information related to the identified DCTN1 variant has been submitted to the ClinVar database (variation ID, 21390; submission ID, 14421829).