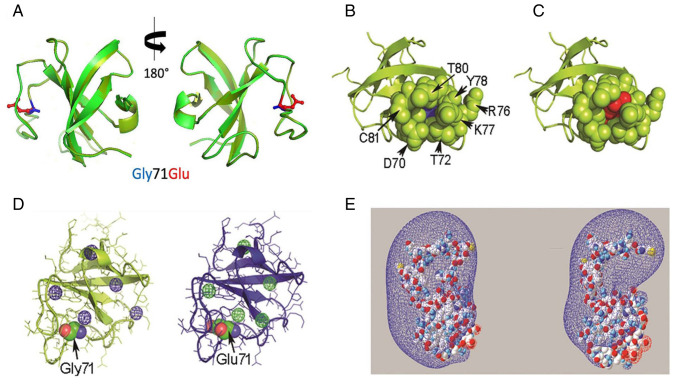
Figure 2.
In silico evaluation of the Gly71Glu variant-spatial relationship of the residue at position 71 of the CAP-Gly domain. (A) Structural alignment of the PDB 2COY template (light green) and Model 1 (dark green). The side chains of Gly71 and Glu71 are displayed in blue and red, respectively. (B) Representative image of residues located spatially close (4 Å) to Gly71 in the wild-type CAP-Gly domain in the free state (PDB 2COY). (C) Equivalent representative image of Glu71 in Model 1. Gly71 and Glu71 are represented as ‘spheres’ in blue and red, respectively. The residues in the proximity of position 71 are shown as green spheres. (D) Different views of the cavities found in PDB 2COY and Model 1 using the Voronoia program suite. The cavities are represented as spheres in a mesh format, in blue or green. (E) Different perspectives of the electrostatic potential map of the PDB 2COY and Model 1 protein structures constructed using Swiss-PdbViewer 4-1-0. The conventional code represents the charge distribution, where blue and red represent the electron-poor and electron-rich regions, respectively. The molecules are represented in a space-filled format, using the CPK color code: Carbon is gray, oxygen is red, nitrogen is blue, sulfur is yellow and hydrogen is white. PDB, protein data bank; CAP-Gly, cytoskeleton-associated protein glycine-rich domain; CPK, Corey-Pauling-Koltun.