ABSTRACT
Tsukamurella, a group of multi-drug resistant, Gram-positive, aerobic, and partially acid-fast bacteria, are emerging causes of bacterial conjunctivitis and keratitis. However, the pathogenesis of Tsukamurella keratitis is largely unknown. To address this, we used New Zealand White rabbits to develop the first eye infection model and conducted in vitro tests to study the pathogenesis mechanisms of Tsukamurella. There is increasing evidence that biofilms play a significant role in ocular infections, leading us to hypothesize that biofilm formation is crucial for effective Tsukamurella infection. In order to look for potential candidate genes which are important in biofilm formation and Tsukamurella keratitis. We performed genome sequencing of two ocular isolates, T. pulmonis-PW1004 and T. tyrosinosolvens-PW899, to identify potential virulence factors. Through in vitro and in vivo studies, we characterized their biological roles in mediating Tsukamurella keratitis. Our findings confirmed that Tsukamurella is an ocular pathogen by fulfilling Koch's postulates, and using genome sequence data, we identified tmytC, encoding a mycolyltransferase, as a crucial gene in biofilm formation and causing Tsukamurella keratitis in the rabbit model. This is the first report demonstrating the novel role of mycolyltransferase in causing ocular infections. Overall, our findings contribute to a better understanding of Tsukamurella pathogenesis and provide a potential target for treatment. Specific inhibitors targeting TmytC could serve as an effective treatment option for Tsukamurella infections.
KEYWORDS: Tsukamurella, mycolyltransferase, virulence, keratitis, conjunctivitis, Koch's postulates
Introduction
Using the information obtained from the analysis of 16S rRNA gene sequences, Tsukamurella was first proposed as a genus in 1988 [1], although the first strain of this group of bacteria was described in 1941 [2], and the first human isolate was reported in 1971 [3]. Similar to related genera of the order Corynebacteriales, such as Nocardia, Rhodococcus, and Gordonia, members of Tsukamurella are Gram-positive, aerobic, catalase-positive, and partially acid-fast as a result of the presence of mycolic acid in the cell envelope. Due to their similar phenotypic properties, differentiation of these genera and speciation within these genera is difficult in most clinical microbiology laboratories [4]. Although over 21 “Tsukamurella species” have been described, using in silico genome-to-genome comparison as the standard of classification, only 16 Tsukamurella species should be included in this genus according to the current state of the taxonomy at the time of writing (https://lpsn.dsmz.de/genus/tsukamurella). Among these 16 species, 12 are known to be associated with human infections. Traditionally, the most commonly reported Tsukamurella infections in humans are indwelling device-related infections, such as catheter-related bacteriemia and peritonitis associated with continuous ambulatory peritoneal dialysis [5–8].
In 2003 and 2009, we first reported Tsukamurella species as novel causes of bacterial conjunctivitis and keratitis respectively [9,10]. Since then, cases of Tsukamurella ocular infections have been noted in other countries [7,11–14]. Similar to keratitis caused by other bacteria, Tsukamurella keratitis is also associated with wearing contact lenses [9]. In the last few years, we have also discovered five additional novel Tsukamurella species, T. sinensis, T. hongkongensis, T. ocularis, T. hominis and T. conjunctivitidis, from patients with ocular infections [15–17]. Moreover, our recent study, based on the largest number of Tsukamurella cases, showed that the number of Tsukamurella isolates recovered from ophthalmological specimens was higher than that recovered from blood cultures [18]. Although ocular infections could be the most important group of disease caused by Tsukamurella, the pathogenesis of Tsukamurella keratitis is largely unknown. We hypothesized that biofilm formation played a key role in Tsukamurella keratitis as well as other indwelling device infections. To test the hypothesis, in the first part of this study, we developed a rabbit model of Tsukamurella keratitis and sequenced and annotated the genomes of T. tyrosinosolvens and T. pulmonis, which have been reported to cause keratitis, in order to identify candidate genes for biofilm formation. In the second part of the study, the candidate genes were studied through gene deletion and complementation experiments as well as in vitro and animal models developed.
Materials and methods
Ethics statement
This study was approved by the institutional review board of The University of Hong Kong (HKU)/Hospital Authority Hong Kong West Cluster in Hong Kong (IRB UW 16-365). The animal experiments were approved by the Committee on the Use of Live Animals in Teaching and Research (CULATR), HKU (CULATR 4794-18 and 4448-17) and the Department of Health, the Government of the HKSAR under the Animals (Control of Experiments) Ordinance, Chapter 340.
Bacterial strains and growth conditions
The source of all Tsukamurella strains used in this study is detailed in Table 1. T. pulmonis-PW1004 was isolated from the serous discharge of a 69-year-old Chinese woman with conjunctivitis, while T. tyrosinosolvens-PW899 was previously isolated from the corneal scraping of an 87-year-old Chinese woman with keratitis (Table 1) [9,10]. Unless otherwise specified, all Tsukamurella isolates were grown on brain heart infusion (BHI) agar at 37°C under aerobic conditions for 48 h (h).
Table 1.
Bacterial strains and plasmids used in this study.
| Strains or plasmids | Genotype and descriptions | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α | F- φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17 (rk-, mk+) gal- phoA supE44 λ- thi-1 gyrA96 relA1 | Invitrogen |
| T. tyrosinosolvens PW899 | Corneal scraping of a patient with keratitis | Woo et al. [9] |
| T. pulmonis PW1004 | Eye swab of a patient with conjunctivitis | Woo et al. [10] |
| PW1004 | PW1004 derivative with galK deletion, ΔgalK | Present study |
| PW1004ΔtmytA | PW1004 derivative with tmytA deletion, ΔgalK | Present study |
| PW1004ΔtmytB | PW1004 derivative with tmytB deletion, ΔgalK | Present study |
| PW1004ΔtmytC | PW1004 derivative with tmytC deletion, ΔgalK | Present study |
| Plasmids | ||
| pCR®-XL-TOPO® | Cloning vector, pUC ori kan | Invitrogen |
| p2NIL | Suicidal plasmid, oriE kan | Parish et al. [46] |
| pNV18 | Tsukamurella/E. coli shuttle vector, aph | Chiba et al. [30] |
| p2NIL-GalK | p2NIL expressing wild-type GalK under the control of the hsp60 promoter, kan | Present study |
| pCR-XL-tmytA | pCR-XL containing tmytA and flanking fragments, kan | Present study |
| pCR-XL-tmytB | pCR-XL containing tmytB and flanking fragments, kan | Present study |
| pCR-XL-tmytC | pCR-XL containing tmytC and flanking fragments, kan | Present study |
| pΔtmytA | p2NIL containing flanking fragments of tmytA, expressing GalK under the control of the hsp60 promoter, kan | Present study |
| pΔtmytB | p2NIL containing flanking fragments of tmytB, expressing GalK under the control of the hsp60 promoter, kan | Present study |
| pΔtmytC | p2NIL containing flanking fragments of tmytC, expressing GalK under the control of the hsp60 promoter, kan | Present study |
| ptmytC | pNV18 expressing ORF of tmytC under the control of the hsp60 promoter, aph, for TmytC protein expression | Present study |
The bacterial cell density was determined by measuring the optical density at 600 nm (OD600). The growth kinetics of the wild-type PW1004 (PW1004-WT), tmytC (Tsukamurella mycolyltransferase C) knockout and complemented mutants were examined by monitoring OD600 at different time points.
Rabbit model for Tsukamurella keratitis
Eight male New Zealand White (NZW) rabbits, 1.5–2.0 kg, were inoculated intrastromally as described previously [19]. Corneas of four rabbits were injected with approximately 10 µl (i.e. 106 CFU) of T. pulmonis-PW1004 using a microliter syringe with a 30G needle. Another four rabbit corneas were challenged with plain culture medium as the control. Rabbits were monitored daily for signs of disease. At 24 h post-infection (PI), rabbit eyes were photographed, evaluated for pathology, and sacrificed. After euthanasia, the corneas of each rabbit were surgically removed for histopathological and immunohistochemical analyses and bacterial enumeration.
Quantification of Tsukamurella cells
To quantify the number of Tsukamurella in the cornea, each tissue was homogenized and diluted in PBS. The diluted sample was plated in triplicate on BHI agar and incubated for 48 h at 37°C. The number of bacteria was expressed as CFU/cornea.
Histopathological and immunohistochemical analyses
Mice antiserum against T. pulmonis-PW1004 was produced by subcutaneously injecting 200 µl of heat-inactivated Tsukamurella cells (i.e. 107 CFU) into three mice, using an equal volume of complete Freund's adjuvant (Sigma, USA) as described previously [20]. Two weeks after the last immunization, 100 µl of blood was collected via the lateral saphenous vein of the mice to obtain the sera. Serum samples collected from a mock-infected mouse were used as the control antibody.
To examine the histopathology of corneal tissues of rabbits challenged with the PW1004-WT and its mutants, excised infected eyes were examined. Corneal tissues were fixed, embedded, and stained with haematoxylin and eosin (H&E) according to Fischer et al. [21]. Histopathological changes were observed using an Olympus BX53 Digital Upright Microscope with DP80 microscope camera (Tokyo, Japan) and imaging system. Immunohistochemical staining for Tsukamurella was performed using anti-T. pulmonis-PW1004 serum using protocols as described previously [20]. Sections were counterstained with haematoxylin. Tissues from sterile saline controls were included as negative controls.
Myeloperoxidase (MPO) assay
MPO is an enzyme used to measure polymorphonuclear leukocyte (PMN) accumulation in tissue samples. The amount of MPO activity in corneal homogenates can indicate the number of infiltrating PMNs. The assay uses a colorimetric reagent, o-dianisidine, to react with the hypochlorite produced by the MPO reaction. One MPO unit of activity is equivalent to approximately 100,000 PMNs [22].
Draft genome sequencing and analysis
The draft genomes of T. tyrosinosolvens-PW899 and T. pulmonis-PW1004 were sequenced using high-throughput sequencing and assembled as previously described [23]. Genomic DNA was extracted from overnight cultures of each Tsukamurella strain grown on BHI agar using the QIAGEN Genomic-tip 20/G kit, following the manufacturer's instructions (QIAGEN). The extracted DNA samples were then subjected to sequencing using the Illumina Hi-Seq 1500 platform, generating 151-bp paired-end reads. The sequence raw reads were processed using Trimmomatic v0.32 to remove adapters, low-quality reads and duplicate reads [24]. De novo assembly of the DNA sequences was performed using Velvet 1.2.10, with default parameters [25]. The resulting sequences were used for the prediction and annotation of protein coding regions. This was performed using version 2.0 of the RAST (Rapid Annotations using Subsystem Technology) server, applying default parameters and the RASTtk annotation scheme, and the Clusters of Orthologous Groups of proteins (COGs) [26,27]. Circular maps were generated using CGview [28]. This Whole Genome Shotgun project has been deposited in DDBJ/EMBL/GenBank under the accession numbers QPKD00000000; BioSample SAMN09691326 for T. tyrosinosolvens-PW899 and QQQF00000000; BioSample SAMN09691327 for T. pulmonis-PW1004.
Construction of non-polar deletion mutant strains
Bacterial strains and plasmids used in this part of the study are listed in Table 1. The wild-type T. pulmonis-PW1004 was a clinical strain isolated from a patient in Hong Kong [10]. Unmarked, non-polar deletion of tmytA, tmytB, and tmytC were constructed respectively by homologous recombination using our newly developed suicide plasmid p2NIL-GalK (Table 1). Primers used for deletion mutagenesis are listed in Supplementary Table 1. The strategy used to generate the tmytC mutant involved amplifying the gene and its upstream and downstream regions from T. pulmonis-PW1004 genomic DNA, purifying the PCR product, and cloning it into pCR-XL-TOPO to generate pCR-XL-tmytC. The 5’ and 3’ flanking regions of tmytC were amplified from plasmid DNA of pCR-XL-tmytC and combined through overlapping PCR to create an in-frame deletion pattern. This pattern was then cloned into p2NIL-GalK, resulting in the final construct pΔtmytC. The construct was introduced into Escherichia coli DH5α by electroporation. Transformants were selected on BHI agar with kanamycin, followed by culturing on BHI agar without antibiotics and selection on BHI agar with 2-deoxy-D-galactose [29]. The resulting 2-deoxy-D-galactose-resistant colonies were assessed for kanamycin sensitivity and screened using primers tmytC-UF-1/DR-1. Similar strategies were used to generate deletion mutants of tmytA and tmytB. Finally, all mutants were confirmed by DNA sequencing with inner-tmytC-F/R primers and the expression level of mycolyltransferase genes in the wild-type (WT) T. pulmonis PW1004 and the three knockout mutants were determined by qRT-PCR.
Complementation of PW1004ΔtmytC
The coding region of tmytC gene, together with its ribosome-binding site, was amplified from chromosomal DNA of PW1004 using primer hsp60-F/R and tmytC-F/R and subcloned into expression shuttle vector pNV18 [30], resulting in the final construct pNV18-tMytC. The complementation plasmid was transformed into PW1004ΔtmytC, and TmytC protein was constitutively expressed. A mutant strain transformed with empty plasmid pNV18 was used as a negative control.
Biofilm growth condition and quantitation
Biofilm was formed in modified M63 medium with slight modifications [31]. Tsukamurella strains cultured in BHI broth at 37°C for 2 days. Saturated bacteria (108 CFU/ml) was then inoculated at 1:1000 in modified M63 medium in a 6-well flat bottom polystyrene plate in triplicate and incubated at 25°C for 14 days without disturbance. Biofilms were quantified using a crystal violet assay. Experiments were performed in triplicate and repeated three times.
Microscopy analysis of the biofilms
For scanning electron microscopy (SEM) analysis, biofilm was fixed in 6% glutaraldehyde for 72 h and observed using LEO 1530 FEC SEM (Zeiss, Germany). Additionally, the biofilm was stained with SYTO 9 green fluorescent nucleic acid stain and visualized using a Perkin Elmer UltraVIEW VOX Spinning Disc confocal laser scanning microscope with 3-dimensional reconstructions created using Volocity 6.3 software.
Hydrophobicity assay
The hydrophobicity assay followed the method by Nguyen et al [32]. Droplets of oil and trypan blue (0.05% solution in sterile water) were applied to the T. pulmonis and its mutant biofilms cultured for 14 days in polystyrene plates. The appearance of the droplets (beading or spreading) was observed and photographed to determine the hydrophobicity of the strains.
Biofilm formation on contact lens and quantification
Senofilcon A soft contact lenses (Johnson & Johnson vision care Inc., Jacksonville, FL, USA) were used in the study. Biofilm quantification followed the method by Szczotka-Flynn et al. with slight modifications [33]. Contact lenses were washed with 1× PBS and incubated in cell suspensions (absorbance of 0.5 at OD600 of the PW1004-WT and its derivative mutants) for 3 days. Lenses were then washed and immersed in 1% TSB for 3 days on a rocking platform. Biofilm was quantified by sonication and vortexing, and the number of CFUs was determined. Experiments were performed in triplicate and repeated three times.
Statistical analysis
Unless stated otherwise, data generated were expressed as mean ± standard error of the mean (SEM) from three independent experiments. Statistical comparison between different groups was performed by the unpaired Student's t-test. An asterisk indicates a significant difference (*, P < 0.05; **, P < 0.01; n.s., not significant).
Results
Rabbit model of Tsukamurella keratitis
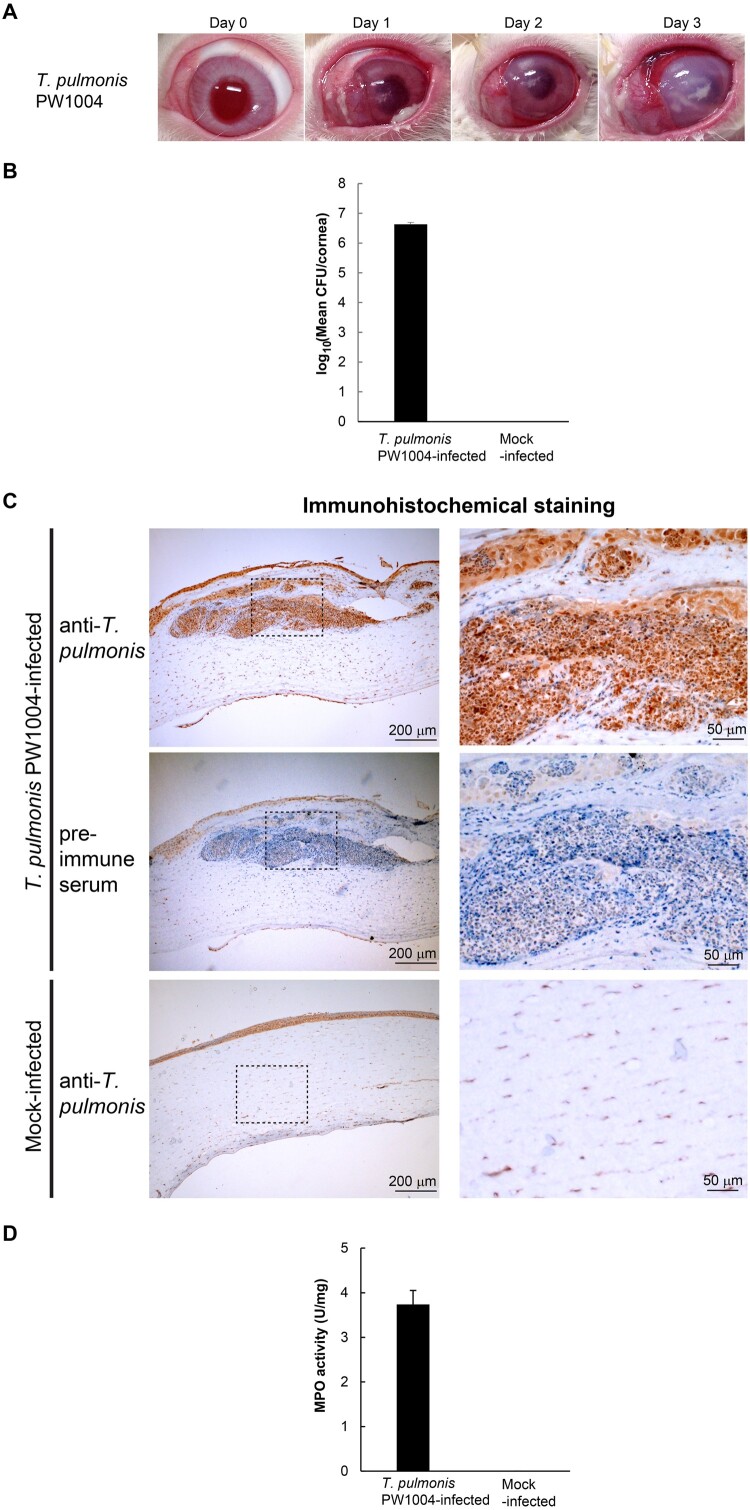
To understand the pathogenesis of Tsukamurella keratitis, we developed an eye infection model using NZW rabbits. We inoculated T. pulmonis-PW1004 at 106 CFUs/cornea by direct intrastromal injection and the pathological changes of the infected rabbit eyes were monitored daily. At 24 h PI, the rabbit corneas started to develop gross pathological signs of infection, including severe iritis, ocular discharge, corneal infiltrate, corneal erosions and dense corneal opacity, with increasing severity observed over time (Figure 1a). On the other hand, we did not observe any pathological signs in control rabbits injected with plain culture medium only (Supplementary Figure 1). At 24 h PI, pure cultures of Tsukamurella at 6.64 ± 0.46 log CFU/cornea were recovered from the infected rabbits (Figure 1b). Active infection was confirmed by the detection of T. pulmonis-PW1004 antigens by immunohistochemical staining using specific anti-T. pulmonis antibodies. Strong staining against T. pulmonis could be detected in corneal sections from rabbits infected with T. pulmonis-PW1004 (top, Figure 1c) but not from the control rabbits (bottom, Figure 1c). Specificity of the anti-T. pulmonis antibodies was verified by staining the corneal sections using mouse pre-immune control serum (middle, Figure 1c). Large amount of inflammatory cell infiltraiton was observed with the haematoxylin counterstain in the corneal sections from the T. pulmonis-infected rabbits (top and middle, Figure 1c). This was in line with the results of the MPO activity assay, which quantified tissue PMN accumulation. MPO activities increased to 3.87 ± 0.26 U/mg in the corneal homogenates from rabbits infected with T. pulmonis-PW1004 compared to those from rabbits of the control group (Figure 1d). To fulfill Koch's postulates, we used T. pulmonis-PW1004 recovered from the cornea of a rabbit to infect other rabbits using the same route of inoculation. Results consistently showed that the infected rabbit developed similar signs and symptoms of keratitis and histopathological changes at 24 h PI.
Figure 1.
Experimentally induced keratitis in NZW rabbits after intrastromal injection of T. pulmonis-PW1004. (a) Gross appearance of the rabbit eyes after Tsukamurella infection. (b) Mean bacterial load recovered from the cornea of rabbits infected with T. pulmonis-PW1004 and those of control rabbits at 24 h PI. Error bars indicated mean CFU/cornea ± SEM of three independent experiments. (c) Immunohistochemical staining of corneal sections using mouse anti-T. pulmonis-PW1004 serum. The boxed area is further enlarged and shown in the right-hand panel of the corresponding image. Strong positive staining in brown colour against T. pulmonis could be detected in corneal sections from rabbits infected with T. pulmonis-PW1004 (top) but not from the mock-infected control rabbits (bottom). The middle panel shows corneal sections from the infected rabbits stained with pre-immune control serum; corneal sections of infected rabbits showing large amount of inflammatory cell infiltration with haematoxylin counterstain (top and middle). (d) MPO activity (U/mg) of the corneal tissues harvested from rabbits. Error bars indicate mean ± SEM of three independent experiments.
Genome sequencing and analysis of T. tyrosinosolvens-PW899 and T. pulmonis-PW1004
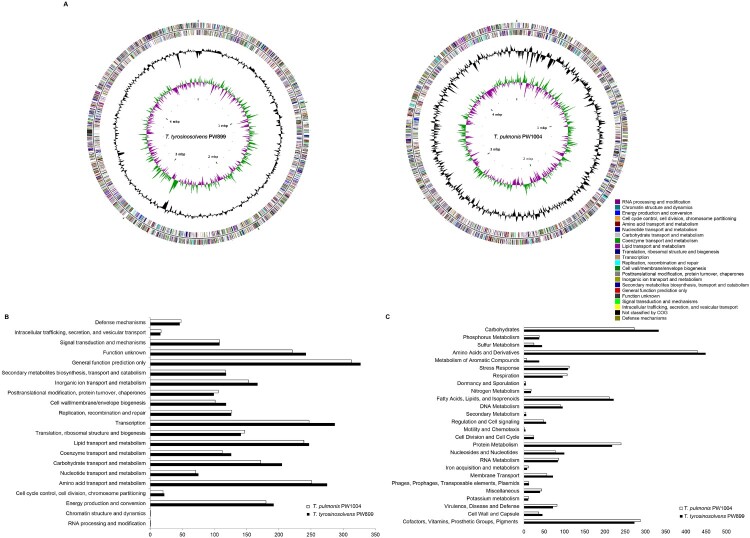
We attempted to sequence Tsukamurella genomes to identify potential virulence factors, particularly those related to biofilm formation, associated with Tsukamurella-mediated ocular infections. The draft genomes of T. tyrosinosolvens-PW899 and T. pulmonis-PW1004, two strains previously isolated from patients with ocular infections [9,10], were sequenced and assembled (Figure 2a). Sequencing generated 11–15 million clean reads per strain (estimated 410-540× coverage). After de novo assembly, the draft genome of T. tyrosinosolvens-PW899 was 4.88 Mb in length distributed in 307 contigs (>500 bp), and that of T. pulmonis-PW1004 was 4.60 Mb in length distributed in 245 contigs (>500 bp) (GenBank accession numbers of T. tyrosinosolvens-PW899 and T. pulmonis-PW1004 are QPKD00000000 and QQQF00000000, respectively) (Table 2). All contigs generated were submitted to the RAST version 2.0 annotation server, resulting in 4558 protein-coding sequences (CDSs), 3 rRNA operons and 49 tRNA-encoding genes for T. tyrosinosolvens-PW899 and 4241 CDSs, and 3 rRNA operons, and 47 tRNA-encoding genes for T. pulmonis-PW1004 (Table 2). Each CDS in the two genomes was further classified into different categories in COGs (Figure 2b) and subsystems in RAST (Figure 2c) based on their predicted functional roles. Specifically, the two Tsukamurella genomes contained a number of putative genes that may be involved in biofilm formation (Supplementary Table 2). Among these genes, three homologues of a gene encoding mycolyltransferase were identified in both genomes of T. tyrosinosolvens-PW899 and T. pulmonis-PW1004. Previous studies have shown that proteins which possess mycolyltransferase activities play significant roles in biofilm formation, as well as other important biological roles, in members of Corynebacteriales [34,35]. For this reason, we attempted to study the role of these homologues and in the pathogenesis of Tsukamurella keratitis.
Figure 2.
Graphical circular maps of the genomes and the distributions of predicted coding sequence function according to COG and SEED subsystems. (a) T. tyrosinosolvens-PW899 (left) T. pulmonis-PW1004 (right). From outside to centre, ring 1 and 2 show protein coding genes on both the forward and reverse strand (coloured by COG categories, respectively); ring 3 shows G + C% content plot, and ring 4 shows GC skew, purple indicating negative values and green, positive values; The columns indicate the number of proteins in different (b) COG and (c) SEED subsystems.
Table 2.
Results of draft genome assembly of T. tyrosinosolvens-PW899 and T. pulmonis-PW1004.
| Genome assembly data | T. tyrosinosolvens-PW899 | T. pulmonis-PW1004 |
|---|---|---|
| Genome size | 4.88 Mb | 4.60 Mb |
| G + C content | 71.0% | 70.9% |
| Total no. of contigs | 321 | 266 |
| No. of contigs (>500 bp) | 307 | 245 |
| No. of predicted protein-coding genes | 4558 | 4241 |
| No. of subsystems | 399 | 384 |
| No. of tRNAs | 49 | 47 |
| No. of rRNA operons | 3 | 3 |
| GenBank accession no. | QPKD00000000 | QQQF00000000 |
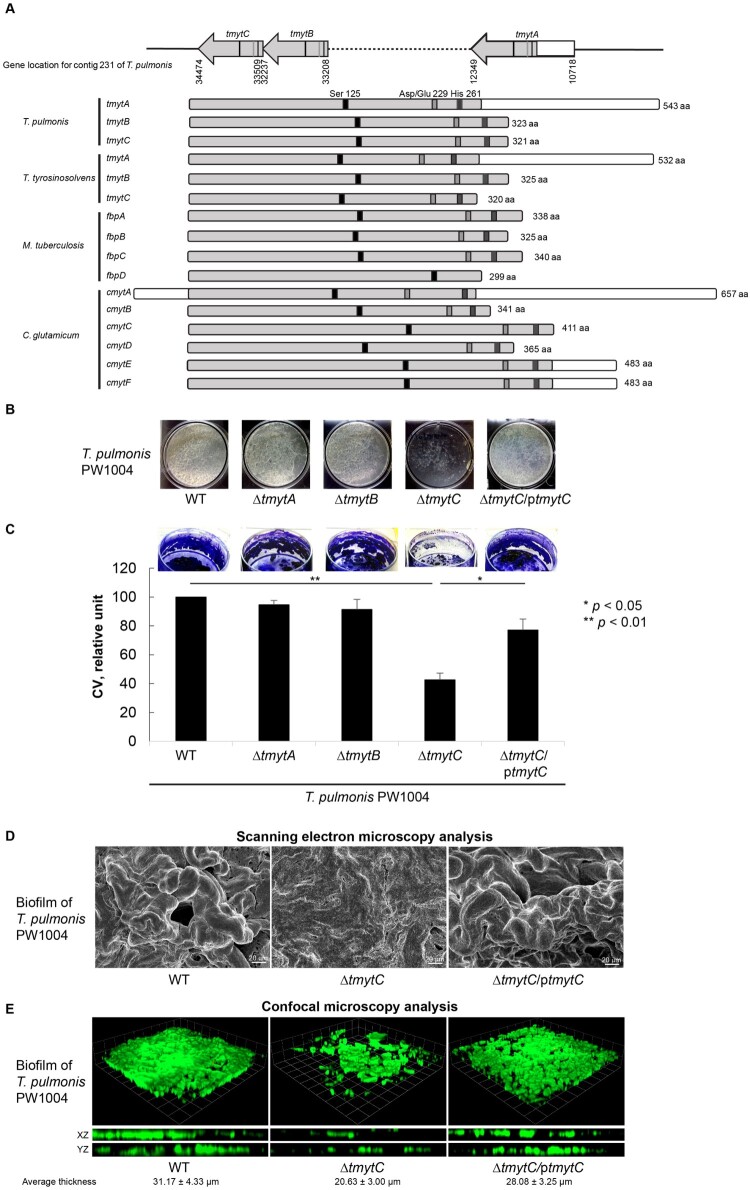
The three mycolyltransferase-encoding homologues identified in both Tsukamurella genomes were designated as tmytA (Tsukamurella mycolyltransferase A), tmytB and tmytC respectively (Figure 3a). The size of the three mycolyltransferase homologues varied from 963 to 1632 bp, as a result of the lack of a C-terminal region in tmytB and tmytC. Their N-terminal regions in T. tyrosinosolvens-PW899 and T. pulmonis-PW1004 shared 38.1–68.9% and 38.9–69.6% amino acid (aa) identities respectively (Figure 3a). Phylogenetically, they were most closely related to the mycolyltransferase of Williamsia limnetica, also a member of Corynebacteriales, sharing 44–45% aa identities. Detailed annotation of the gene sequences revealed the presence of critical aa residues Ser-125, Asp/Glu-229, and His-261 in their N-terminal regions, which forms a catalytic triad (Figure 3a). This is a typical feature of other characterized mycolyltransferases and is essential for mycolyltransferase activity [35]. To further characterize the functional role of the mycolyltransferase homologues in Tsukamurella, individual knockout mutant strains (PW1004ΔtmytA, PW1004ΔtmytB, and PW1004ΔtmytC) were constructed respectively in T. pulmonis-PW1004. Biofilm phenotypes in vitro and virulence in vivo of each mutant strain were studied and compared to those of wild-type and/or complemented strains.
Figure 3.
Characterization of the 3 tmyt homologues in Tsukamurella. (a) Locations of the tmytA, tmytB, and tmytC gene in the genome of T. pulmonis-PW1004 are indicated. Alignment of the tmyt homologues identified in T. pulmonis-PW1004, T. tyrosinosolvens-PW899, M. tuberculosis (GenBank accession numbers NP_218321, NP_216402, YP_177694 and YP_178017) and C. glutamicum (GenBank accession numbers AAAP23202-AAAP232007). The catalytic triad formed by functional residues Ser125, Asp/Glu229, and His261, which are important for mycolyltransferase activity, are indicated by black, grey, and dark grey boxes, respectively. (b) Biofilm formed by the PW1004-WT and its derivative mutants when they were cultured under static conditions for 3 days. With the exception of the PW1004ΔtmytC, dense and confluent biofilm was formed as a floating pellicle at the air-liquid interface in PW1004ΔtmytA, PW1004ΔtmytB, PW1004-WT, and tmytC complemented mutant (PW1004ΔtmytC/ptmytC). (c) Quantitation of the biofilm formed by the PW1004-WT and its derivative mutants using crystal violet staining method. The amount of biofilms was significantly reduced in PW1004ΔtmytC compared to the PW1004-WT (P < 0.01) and complemented mutant (P < 0.05). (d) SEM and (e) confocal microscopy analyses of Tsukamurella biofilms cultured under static conditions for 3 days. Representative SEM micrographs of the biofilm formed by the PW1004ΔtmytC was flattened and less structured compared to those formed by the PW1004-WT and complemented mutant. Likewise, biofilms were fixed and stained with SYTO 9 green fluorescent stain prior to confocal microscopy analysis. Representative micrographs comparing biofilm thickness of each Tsukamurella strain was measured in different points of each field. The means and standard deviations of three independent experiments are shown.
Characterization of biofilm phenotypes in wild-type PW1004 and the 3 tmyt knockout mutants
We successfully constructed non-polar deletions of tmytA, tmytB, and tmytC genes in T. pulmonis-PW1004 and confirmed them through DNA sequencing and gene expression analysis using qRT-PCR. We hypothesized that knockout of the tymt gene may impair biofilm formation in Tsukamurella. To investigate this, the wild-type and 3 tymt knockout mutants of T. pulmonis-PW1004 were cultured under static conditions to allow the formation of biofilms. Results showed that dense and confluent biofilms were formed as a floating pellicle at the air–liquid interface in PW1004ΔtmytA, PW1004ΔtmytB, and PW1004-WT, whereas less textured and reticulated biofilms were formed in PW1004ΔtmytC (Figure 3b). Biofilm quantification using the crystal violet staining method showed that the amount of biofilm was significantly reduced in PW1004ΔtmytC compared to PW1004-WT (P < 0.01), while PW1004ΔtmytA and PW1004ΔtmytB produced similar levels of biofilm as PW1004-WT (Figure 3c). To rule out the possibility that knockout of tmyt genes may alter growth kinetics and hence biofilm formation, we measured the growth kinetics of the wild-type and knockout mutants and found that there was no significant difference between the growth rates of the wild-type and knockout mutants (Supplementary Figure 2). To confirm the importance of TmytC in T. pulmonis biofilm formation, we transformed a TmytC expression plasmid (pNV18-tmytC) into PW1004ΔtmytC and studied the subsequent biofilm characteristics. Results showed that PW1004ΔtmytC complemented with TmytC (PW1004ΔtmytC/ptmytC) restored the biofilm phenotype with biofilm quantity levels similar to that of PW1004-WT (Figure 3b, c). Independently, we performed SEM analysis to study the biofilm structure of WT, tmytC-knockout, and complemented PW1004. Results showed that the biofilm formed by PW1004-WT was highly structured (left, Figure 3d). Deletion of the tmytC gene, however, resulted in the formation of flatter and less structured biofilms (middle, Figure 3d), whereas PW1004ΔtmytC/ptmytC complemented mutant displayed similar biofilm phenotypes as PW1004-WT (right, Figure 3d). The results were further supported by confocal microscopy analysis, in which optical sectioning along the Z axis showed reduced biofilm formation with an average thickness of 20.63 ± 3.00 µm in PW1004ΔtmytC compared to those of 31.17 ± 4.33 µm and 28.08 ± 3.25 µm in the wild-type and complemented mutant, respectively (P < 0.001), with no significant difference in the thickness of the biofilm formed between the wild-type and complemented mutant (Figure 3e). Collectively, the results supported that deletion of tmytC gene, but not tmytA or tmytB genes, perturbed the biofilm-forming capacity of T. pulmonis-PW1004, suggesting that mycolyltransferase contributes significantly to biofilm formation in T. pulmonis.
Reduced virulence in tmytC knockout mutant in rabbits
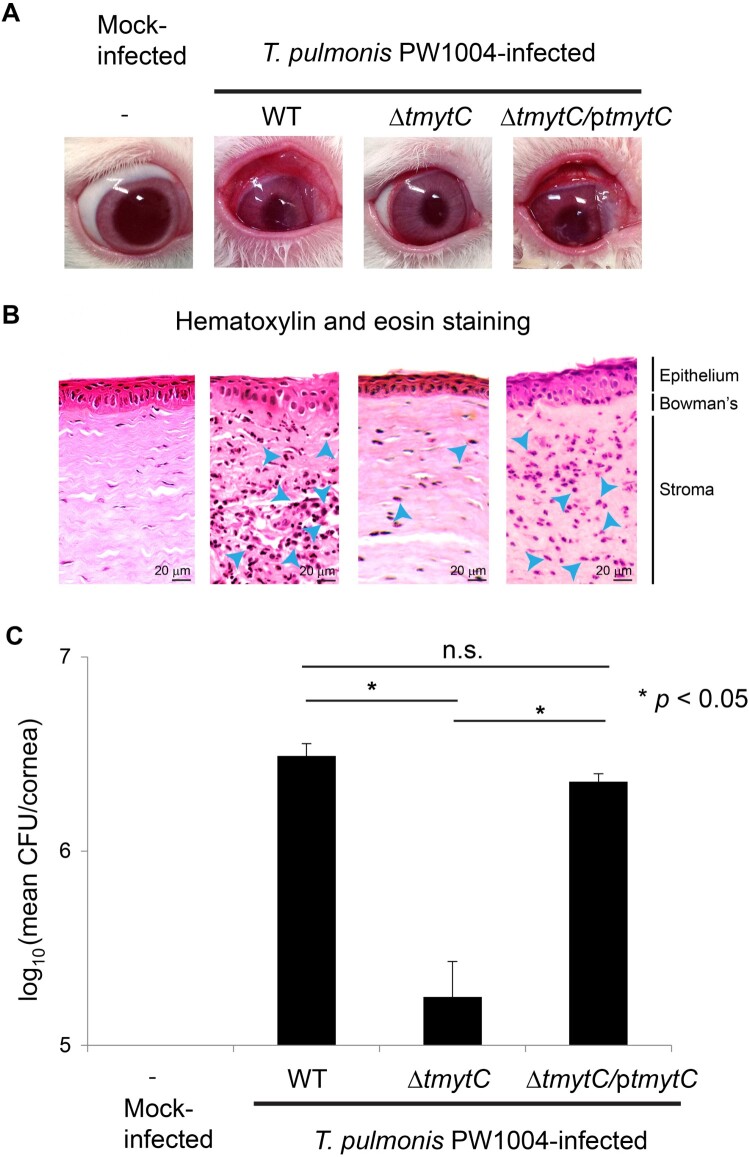
We hypothesized that biofilm formation is one of the major factors contributing to bacterial virulence in Tsukamurella keratitis. Hence, we examined the importance of TmytC in the pathogenesis of Tsukamurella keratitis in vivo by inoculating rabbit corneas with PW1004-WT, PW1004ΔtmytC, and PW1004ΔtmytC/ptmytC respectively via intrastromal injection. The keratitis symptoms in rabbits infected with PW1004ΔtmytC were less severe with less purulent discharge compared to those infected with PW1004-WT and PW1004ΔtmytC/ptmytC (Figure 4a). On day 2 PI, the rabbits were sacrificed and their corneas were harvested for histopathological studies and bacterial counts. Consistent with the gross appearance of the eyes (Figure 4a), histopathological examination of the corneal tissues revealed a lower degree of PMN infiltration in rabbits infected with PW1004ΔtmytC as compared to PW1004-WT and PW1004ΔtmytC/ptmytC, where PMN infiltration was apparent in corneal stroma and stromal oedema was prominent (Figure 4b). In addition, PW1004-WT and PW1004ΔtmytC/ptmytC showed a disordered arrangement of the epithelial layer with focal loss of superficial epithelial cells, and shrinkage of the Bowman's membrane (Figure 4b). Such damages were minimally apparent in PW1004ΔtmytC and absent in the normal corneal sections (Figure 4b). Moreover, the corneal bacterial loads of PW1004-WT and PW1004ΔtmytC/ptmytC were significantly higher than that of PW1004ΔtmytC (P < 0.05), which showed only about 5% recovery rate on day 2 PI (Figure 4c). Taken together, these results suggested that TmytC contributes to the pathogenesis of Tsukamurella keratitis, probably through enhancing the adherence of Tsukamurella to corneal epithelial cells in vivo.
Figure 4.
TmytC is a virulence factor of T. pulmonis. (a) Gross appearance of the rabbit eyes after intrastromal injection of the PW1004-WT and its derivative mutants. The symptoms of keratitis in rabbits infected with PW1004ΔtmytC appeared less severe with fewer purulent discharges compared to those infected with the PW1004-WT and PW1004ΔtmytC/ptmytC mutant. (b) Representative images of H&E staining of corneal sections of infected rabbits. PMN (shown in blue arrows) infiltration in corneal stroma and stromal oedema was less prominent in PW1004ΔtmytC compared to the PW1004-WT and PW1004ΔtmytC/ptmytC mutants, showing the disorderly arranged epithelial layer. (c) Mean bacterial load in cornea inoculated with the PW1004-WT, PW1004ΔtmytC, and PW1004ΔtmytC/ptmytC at day 2 PI. Error bars indicate means ± SEM of three independent experiments.
Reduced bacterial adhesion on contact lens in tmytC knockout mutant
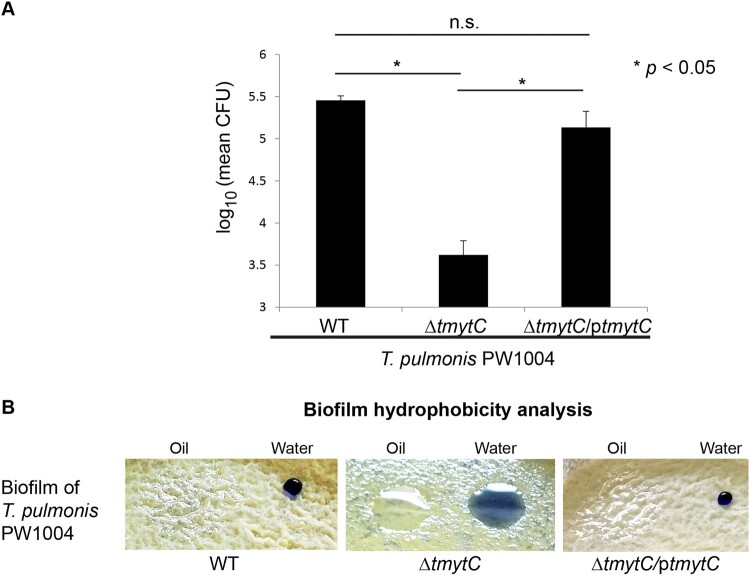
Adhesion ability is a key characteristic related to biofilm formation [36]. Since there was an apparent change in the biofilm phenotypes and impaired biofilm formation capacity of PW1004ΔtmytC (Figure 3b-e), we hypothesized that Tsukamurella adherence to abiotic materials, such as contact lenses, may also be altered. Therefore, the adhesion ability of PW1004-WT, PW1004ΔtmytC and PW1004ΔtmytC/ptmytC to contact lenses was determined quantitatively. Results showed that initially, all three strains were able to form a thin layer of cells over the entire surface of the contact lens. However, after subsequent mechanical detachment, washing and re-culturing, the number of bacteria recovered from the contact lenses inoculated with PW1004ΔtmytC (3.62 ± 0.17 log CFU) was significantly lower compared to those inoculated with PW1004-WT (5.46 ± 0.05 log CFU) and PW1004ΔtmytC/ptmytC (5.13 ± 0.19 log CFU), respectively (P < 0.05) (Figure 5a). Consistently, during the culturing process of the three bacterial strains, a ring of cells adhering to the air–liquid interface was observed in the culture tubes of PW1004-WT and PW1004ΔtmytC/ptmytC, but not PW1004ΔtmytC (Supplementary Figure 3). In contrast to PW1004-WT and PW1004ΔtmytC/ptmytC, a large cell pellet was observed at the bottom of the culture tube of PW1004ΔtmytC (Supplementary Figure 3), implicating its weaker adhesion to plastic tubes. Collectively, these results demonstrated that the tmytC knockout mutant possessed reduced adhesion ability to contact lenses and abiotic surface (i.e. plastic culture tube), suggesting that TmytC played a role in the adherence of Tsukamurella to abiotic surfaces in vitro.
Figure 5.
Altered adherence to contact lens, biofilm hydrophobicity, and PHMB susceptibility of the tmytC knockout mutant. (a) Adherence of the wild-type and tymtC mutants to contact lenses in vitro as analysed by CFU counting analysis. The number of bacteria recovered from the contact lenses inoculated with PW1004ΔtmytC was significantly lower compared to those inoculated with the PW1004-WT (P < 0.05) and the PW1004ΔtmytC/ptmytC (P < 0.05) mutants. Error bars indicated means ± SEM of three independent experiments. (b) The PW1004ΔtmytC surface was more hydrophilic compared to the PW1004-WT. Droplets of oil or water containing trypan blue were applied to the surface of the biofilm lawn. In the PW1004-WT and tymtC complemented mutant, the oil spread into a thin film over the surface, suggesting that the cell surface was hydrophobic in nature. In contrast, the water droplet continued to spread (i.e. more hydrophilic) and the oil droplet spread less (i.e. less hydrophobic) in the tymtC knockout mutant.
Decreased cell surface hydrophobicity in the tmytC knockout mutant
In addition to adhesion ability, cell surface hydrophobicity is another characteristic related to biofilm formation [37]. The reduced adhesion ability previously observed for the PW1004ΔtmytC mutant (Figure 5a; Supplementary Figure 3) prompted us to study the cell surface hydrophobicity in the wild-type and mutants using a visualization experiment as described previously [32]. This experiment involves adding a drop of water or oil onto the biofilm culture. If the culture cell surface is hydrophobic in nature, the water will form a bead and the oil will spread over the surface, as observed for the wild-type strain (left, Figure 5b). On the other hand, altered cell surface hydrophobicity was clearly shown in the PW1004ΔtmytC biofilm, in which the drop of water spread out more (i.e. more hydrophilic), while oil spread out less (i.e. less hydrophobic) (middle, Figure 5b), compared to the wild-type. The biofilm hydrophobicity of the complemented mutant was restored to a comparable level as observed in the wild-type (right, Figure 5b). These observations indicated that TmytC also contributed to the cell surface hydrophobicity of T. pulmonis.
Discussion
In this study, we documented that Tsukamurella is an ocular pathogen by fulfilling Koch's postulates using the NZW rabbit keratitis model. Bacterial keratitis is an ophthalmologic emergency that requires prompt diagnosis and expedient treatment so as to prevent visual loss. It is often associated with wearing contact lenses or other microtrauma, such as trichiasis, to the epithelial surface of the cornea. Among the cases of bacterial keratitis of which an aetiology can be identified, most are caused by Staphylococcus aureus and Streptococcus pneumoniae. Similar to animal studies in S. aureus [38], we demonstrated that the NZW rabbit is an excellent model for Tsukamurella keratitis. T. pulmonis-PW1004 was recovered in abundance and in pure culture from all rabbits infected with Tsukamurella, but the bacteria were not recovered in the control group (Figure 1b). The rabbits with keratitis caused by T. pulmonis-PW1004 produced clinical features similar to those observed in patients with ocular infections (Figure 1a). Histological examination further revealed marked pathological damage in corneal tissues of Tsukamurella inoculated rabbits but not in control rabbits (Figures 1c, 4b). When isolates of T. pulmonis recovered from rabbits were used to infect another group of healthy rabbits, it caused keratitis with the same clinical, pathological and histopathological characteristics, fulfilling the Koch's postulate. This animal model was used for the downstream pathogenesis studies.
tmytC, which encodes mycolyltransferase, is important for biofilm formation in T. pulmonis. Bacterial biofilms constitute a unique shield against antibiotic treatment and host immune reactions and is a crucial protective mechanism for bacteria that cause indwelling device infections [39]. As there is increasing evidence that biofilms play a significant role in ocular infections [40] and these as well as indwelling device infections constitute more than 90% of infections caused by Tsukamurella, we hypothesized that this group of bacteria is capable of producing biofilms. In this study, genome sequencing and annotation of the two ocular isolates, T. pulmonis-PW1004 and T. tyrosinosolvens-PW899, revealed a number of genes that may be involved in biofilm formation (Supplementary Table 2). Among these genes, both genomes contained three homologues of genes (tmytA, tmytB, and tmytC) encoding mycolyltransferase (Figure 3a). This enzyme functions by transferring one mycolate residue from trehalose monomycolate (TMM) to another molecule of TMM yielding trehalose 6,6’-dimycolate, which will then be processed by esterase to form free mycolic acids [41]. Mycolic acids contribute to many important biological roles in bacteria, such as being a key component of biofilms [31,34,35], maintaining cell wall structure [32], and mediating host cell immune activation [42]. A close relationship between mycolic acids and biofilm production has been reported in previous studies [34,35]. For example, impaired biofilm formation due to the deletion of a gene encoding mycolyltransferase has been reported in Mycobactereium smegmatis [35]. In this bacterium, five mycolyltransferase gene homologues were identified in its genome. Deletion of fbpA, but not the other four homologues, showed reduced biofilm formation and level of mycolic acids [35]. Unlike M. smegmatis, which possesses five mycolyltransferase gene homologues, we only identified three mycolyltransferase homologues in each of the T. pulmonis-PW1004 and T. tyrosinosolvens-PW899 genomes. The size of these three mycolyltransferase homologues varied considerably due to the lack of a C-terminal region in two of them, similar to the situation in Corynebacterium glutamicum [43] (Figure 3a). in this study, we showed that deletion of tmytC, but not tmytA or tmytB, showed altered biofilm phenotype and impaired biofilm formation in T. pulmonis-PW1004, which was probably due to reduced mycolic acid production (Figure 3b–e). This shows that tmytC is important for biofilm formation in T. pulmonis. However, it remains unclear why disruption of tmytC, but not tmytA or tmytB in Tsukamurella, showed impaired biofilm production, although the 3 mycolyltransferase homologues possess identical essential motifs in their N-terminus (Figure 3a). It is speculated that mycolyltransferase homologues may exhibit differential substrate preference and/or spatial cellular localizations resulting in different activities [32]. Given the important biological roles of mycolic acids, further studies on deciphering the types, chemistry and structure of mycolic acids synthesized in Tsukamurella, and their specific roles in biofilm formation are of crucial importance to understand this emerging ocular pathogen.
TmytC is a virulence factor of Tsukamurella. Since tmytC is crucial for biofilm formation, we further examined its importance for the virulence of T. pulmonis using the NZW rabbit keratitis model. Our results showed that there was marked attenuation of virulence and survival when tmytC was deleted in T. pulmonis-PW1004, but these phenotypes were restored when the gene was complemented (Figure 4a–c). Furthermore, our subsequent mechanistic studies have also revealed that tmytC knockout mutant exhibited impaired adherence to contact lens in vitro (Figure 5a), and that deletion of tmytC led to decreased hydrophobicity in T. pulmonis-PW1004 biofilms (Figure 5b), in line with our previous observation that T. pulmonis was associated with keratitis in a contact lens wearer [9]. In general, bacteria with greater surface hydrophobicity adhere to various contact lenses in greater numbers than hydrophilic bacteria [44,45]. This has been demonstrated in P. aeruginosa, which is known to be highly hydrophobic, exhibited significantly greater adhesion to most contact lens types compared to other ocular pathogens that are less hydrophobic, such as S. aureus, S. pneumoniae, and Haemophilus influenzae [44,45]. This phenomenon is in line with our present results, in which the tmytC knockout mutant with decreased hydrophobicity also showed lower adherence to contact lenses compared to PW1004-WT and the complemented mutant (Figure 5a, b). Further sequence anlayses revealed that TmytC is present in all Tsukamurella species for which genome sequences are available, suggesting that TmytC is highly prevalent in Tsukamurella sepcies and likely plays a crucial role in biofilm formation and virlence in these species. Taken together, the present results supported that TmytC is an important virulence factor for Tsukamurella keratitis, which is mediated through enhancing its adherence to abiotic (i.e. contact lenses, plastics) and biotic (i.e. corneal epithelial cells) surfaces and forming biofilm. Further studies on the development of inhibitors targeting TmytC are warranted for the treatment of Tsukamurella infections.
Supplementary Material
Acknowledgements
We thank the members of the Centre for Genomic Sciences at The University of Hong Kong for their technical support.
Funding Statement
This work is partly supported by the Feature Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE-113-S-0023-A) in Taiwan; and the donation of TE Health Consultant Company Limited.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Author contributions
JT conceived and designed the study and contributed reagents, performed the laboratory and animal work, analysed and interpreted results and wrote the manuscript. YT performed the laboratory and animal work, analysed and interpreted results. SW contributed reagents, analysed and interpreted results. MY, CC, EC, and JF performed the laboratory work, analysed and interpreted results. JC and LX performed the animal work. RAY performed histological analyses. SL conceived and designed the study and contributed reagents, analysed and interpreted results. PW conceived and designed the study and contributed reagents, analysed and interpreted results and wrote the manuscript.
Data/code availability
This Whole Genome Shotgun project has been deposited in DDBJ/EMBL/GenBank under the accession numbers QPKD00000000; BioSample SAMN09691326 for T. tyrosinosolvens-PW899 and QQQF00000000; BioSample SAMN09691327 for T. pulmonis-PW1004.
References
- 1.Collins MD, Smida J, Dorsch M, et al. . Tsukamurella gen. nov. harboring corynebacterium paurometabolum and Rhodococcus aurantiacus. Int J Syst Evol Microbiol. 1988;38(4):385–391. [Google Scholar]
- 2.Steinhaus EA. A study of the bacteria associated with thirty species of insects. J Bacteriol. 1941;42(6):757–790. doi: 10.1128/jb.42.6.757-790.1941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tsukamura M, Mizuno S.. [A new species Gordona aurantiaca occurring in sputa of patients with pulmonary disease]. Kekkaku: [tuberculosis]. 1971 Apr;46(4):93–98. [PubMed] [Google Scholar]
- 4.Teng JL, Tang Y, Chiu TH, et al. . The groEL gene is a promising target for species-level identification of Tsukamurella. J Clin Microbiol. 2017 Feb;55(2):649–653. doi: 10.1128/JCM.02260-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouza E, Perez-Parra A, Rosal M, et al. . Tsukamurella: a cause of catheter-related bloodstream infections. Eur J Clin Microbiol Infect Dis. 2009 Feb;28(2):203–210. doi: 10.1007/s10096-008-0607-2 [DOI] [PubMed] [Google Scholar]
- 6.Schwartz MA, Tabet SR, Collier AC, et al. . Central venous catheter-related bacteremia due to Tsukamurella species in the immunocompromised host: a case series and review of the literature. Clin Infect Dis. 2002 Oct 1;35(7):e72–e77. doi: 10.1086/342561 [DOI] [PubMed] [Google Scholar]
- 7.Liu CY, Lai CC, Lee MR, et al. . Clinical characteristics of infections caused by Tsukamurella spp. and antimicrobial susceptibilities of the isolates. Int J Antimicrob Agents. 2011 Dec;38(6):534–537. doi: 10.1016/j.ijantimicag.2011.07.018 [DOI] [PubMed] [Google Scholar]
- 8.Shaer AJ, Gadegbeku CA.. Tsukamurella peritonitis associated with continuous ambulatory peritoneal dialysis. Clin Nephrol. 2001 Sep;56(3):241–246. [PubMed] [Google Scholar]
- 9.Woo PC, Fong AH, Ngan AH, et al. . First report of Tsukamurella keratitis: association between T. tyrosinosolvens and T. pulmonis and ophthalmologic infections. J Clin Microbiol. 2009 Jun;47(6):1953–1956. doi: 10.1128/JCM.00424-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Woo PC, Ngan AH, Lau SK, et al. . Tsukamurella conjunctivitis: a novel clinical syndrome. J Clin Microbiol. 2003 Jul;41(7):3368–3371. doi: 10.1128/JCM.41.7.3368-3371.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Almeida DR, Miller D, Alfonso EC.. Tsukamurella: an emerging opportunistic ocular pathogen. Can J Ophthalmol. 2010 Jun;45(3):290–293. doi: 10.3129/i09-252 [DOI] [PubMed] [Google Scholar]
- 12.Tam PM, Young AL, Cheng L, et al. . Tsukamurella: an unrecognized mimic of atypical mycobacterial keratitis? The first case report. Cornea. 2010 Mar;29(3):362–364. doi: 10.1097/ICO.0b013e3181ae2594 [DOI] [PubMed] [Google Scholar]
- 13.Park BJ, Goosey JD, Belloso M.. Tsukamurella keratitis: the first case in the United States. Can J Ophthalmol. 2021 Oct;56(5):e153–e155. doi: 10.1016/j.jcjo.2021.03.005 [DOI] [PubMed] [Google Scholar]
- 14.Leung KCP, Au SCL, Ko TCS.. Ophthalmic manifestation of Tsukamurella species: A case series and first report of ocular implant infection after enucleation. Cornea. 2019 Oct;38(10):1328–1331. doi: 10.1097/ICO.0000000000001997 [DOI] [PubMed] [Google Scholar]
- 15.Teng JL, Tang Y, Wong SS, et al. . Tsukamurella hongkongensis sp. nov. and tsukamurella sinensis sp. nov., isolated from patients with keratitis, catheter-related bacteraemia and conjunctivitis. Int J Syst Evol Microbiol. 2016 Jan;66(1):391–397. doi: 10.1099/ijsem.0.000733 [DOI] [PubMed] [Google Scholar]
- 16.Teng JLL, Tang Y, Wong SSY, et al. . Tsukamurella ocularis sp. nov. and tsukamurella hominis sp. nov., isolated from patients with conjunctivitis in Hong Kong. Int J Syst Evol Microbiol. 2018 Mar;68(3):810–818. doi: 10.1099/ijsem.0.002589 [DOI] [PubMed] [Google Scholar]
- 17.Teng JLL, Fong JYH, Fok KMN, et al. . Tsukamurella asaccharolytica sp. nov. Tsukamurella conjunctivitidis sp. nov. and Tsukamurella sputi sp. nov., isolated from patients with bacteraemia, conjunctivitis and respiratory infection in Hong Kong. Int J Syst Evol Microbiol. 2020 Feb;70(2):995–1006. [DOI] [PubMed] [Google Scholar]
- 18.Teng JLL, Tang Y, Wong SSY, et al. . MALDI-TOF MS for identification of tsukamurella species: tsukamurella tyrosinosolvens as the predominant species associated with ocular infections. Emerg Microbes Infect. 2018 May 9;7(1):80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanders ME, Norcross EW, Moore QC, 3rd, et al. . Efficacy of besifloxacin in a rabbit model of methicillin-resistant Staphylococcus aureus keratitis. Cornea. 2009 Oct;28(9):1055–1060. [DOI] [PubMed] [Google Scholar]
- 20.Lau SK, Woo PC, Li KS, et al. . Identification of novel rosavirus species that infects diverse rodent species and causes multisystemic dissemination in mouse model. PLoS Pathog. 2016 Oct;12(10):e1005911. doi: 10.1371/journal.ppat.1005911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fischer AH, Jacobson KA, Rose J, et al. . Hematoxylin and eosin staining of tissue and cell sections. CSH Protoc. 2008 May 1;2008:pdb prot4986. [DOI] [PubMed] [Google Scholar]
- 22.Girgis DO, Dajcs JJ, O’Callaghan RJ.. Phospholipase A2 activity in normal and Staphylococcus aureus-infected rabbit eyes. Invest Ophthalmol Visual Sci. 2003 Jan;44(1):197–202. doi: 10.1167/iovs.02-0548 [DOI] [PubMed] [Google Scholar]
- 23.Teng JL, Tang Y, Huang Y, et al. . Phylogenomic analyses and reclassification of species within the genus Tsukamurella: insights to species definition in the post-genomic Era. Front Microbiol. 2016;7:1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bolger AM, Lohse M, Usadel B.. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014 Aug 1;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zerbino DR, Birney E.. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008 May;18(5):821–829. doi: 10.1101/gr.074492.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Overbeek R, Olson R, Pusch GD, et al. . The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 2014 Jan;42(Database issue):D206-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Galperin MY, Makarova KS, Wolf YI, et al. . Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015 Jan;43(Database issue):D261-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stothard P, Wishart DS.. Circular genome visualization and exploration using CGView. Bioinformatics. 2005 Feb 15;21(4):537–539. doi: 10.1093/bioinformatics/bti054 [DOI] [PubMed] [Google Scholar]
- 29.Mishra A, Wu C, Yang J, et al. . The actinomyces oris type 2 fimbrial shaft FimA mediates co-aggregation with oral streptococci, adherence to red blood cells and biofilm development. Mol Microbiol. 2010 Aug;77(4):841–854. doi: 10.1111/j.1365-2958.2010.07252.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chiba K, Hoshino Y, Ishino K, et al. . Construction of a pair of practical Nocardia-Escherichia coli shuttle vectors. Jpn J Infect Dis. 2007 Feb;60(1):45–47. doi: 10.7883/yoken.JJID.2007.45 [DOI] [PubMed] [Google Scholar]
- 31.Ojha A, Anand M, Bhatt A, et al. . GroEL1: a dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell. 2005 Dec 2;123(5):861–873. doi: 10.1016/j.cell.2005.09.012 [DOI] [PubMed] [Google Scholar]
- 32.Nguyen L, Chinnapapagari S, Thompson CJ.. FbpA-dependent biosynthesis of trehalose dimycolate is required for the intrinsic multidrug resistance, cell wall structure, and colonial morphology of Mycobacterium smegmatis. J Bacteriol. 2005 Oct;187(19):6603–6611. doi: 10.1128/JB.187.19.6603-6611.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Szczotka-Flynn LB, Imamura Y, Chandra J, et al. . Increased resistance of contact lens-related bacterial biofilms to antimicrobial activity of soft contact lens care solutions. Cornea. 2009 Sep;28(8):918–926. doi: 10.1097/ICO.0b013e3181a81835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ojha AK, Baughn AD, Sambandan D, et al. . Growth of Mycobacterium tuberculosis biofilms containing free mycolic acids and harbouring drug-tolerant bacteria. Mol Microbiol. 2008 Jul;69(1):164–174. doi: 10.1111/j.1365-2958.2008.06274.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ojha AK, Trivelli X, Guerardel Y, et al. . Enzymatic hydrolysis of trehalose dimycolate releases free mycolic acids during mycobacterial growth in biofilms. J Biol Chem. 2010 Jun 4;285(23):17380–9. doi: 10.1074/jbc.M110.112813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stoodley P, Sauer K, Davies DG, et al. . Biofilms as complex differentiated communities. Annu Rev Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705 [DOI] [PubMed] [Google Scholar]
- 37.Krasowska A, Sigler K.. How microorganisms use hydrophobicity and what does this mean for human needs? Front Cell Infect Microbiol. 2014;4:112. doi: 10.3389/fcimb.2014.00112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marquart ME. Animal models of bacterial keratitis. J Biomed Biotechnol. 2011;2011:680642. doi: 10.1155/2011/680642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Caldara M, Belgiovine C, Secchi E, et al. . Environmental, microbiological, and immunological features of bacterial biofilms associated with implanted medical devices. Clin Microbiol Rev. 2022 Apr 20;35(2):e0022120. doi: 10.1128/cmr.00221-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elder MJ, Stapleton F, Evans E, et al. . Biofilm-related infections in ophthalmology. Eye. 1995;9(Pt 1):102–109. doi: 10.1038/eye.1995.16 [DOI] [PubMed] [Google Scholar]
- 41.Belisle JT, Vissa VD, Sievert T, et al. . Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science. 1997 May 30;276(5317):1420–1422. doi: 10.1126/science.276.5317.1420 [DOI] [PubMed] [Google Scholar]
- 42.Marrakchi H, Laneelle MA, Daffe M.. Mycolic acids: structures, biosynthesis, and beyond. Chem Biol. 2014 Jan 16;21(1):67–85. doi: 10.1016/j.chembiol.2013.11.011 [DOI] [PubMed] [Google Scholar]
- 43.Ramulu HG, Adindla S, Guruprasad L.. Analysis and modeling of mycolyl-transferases in the CMN group. Bioinformation. 2006 Jun 18;1(5):161–169. doi: 10.6026/97320630001161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bruinsma GM, van der Mei HC, Busscher HJ.. Bacterial adhesion to surface hydrophilic and hydrophobic contact lenses. Biomaterials. 2001 Dec;22(24):3217–3224. doi: 10.1016/S0142-9612(01)00159-4 [DOI] [PubMed] [Google Scholar]
- 45.Dutta D, Cole N, Willcox M.. Factors influencing bacterial adhesion to contact lenses. Mol Vision. 2012;18:14–21. [PMC free article] [PubMed] [Google Scholar]
- 46.Parish T, Stoker NG.. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology. 2000 Aug;146(Pt 8):1969–1975. doi: 10.1099/00221287-146-8-1969 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This Whole Genome Shotgun project has been deposited in DDBJ/EMBL/GenBank under the accession numbers QPKD00000000; BioSample SAMN09691326 for T. tyrosinosolvens-PW899 and QQQF00000000; BioSample SAMN09691327 for T. pulmonis-PW1004.