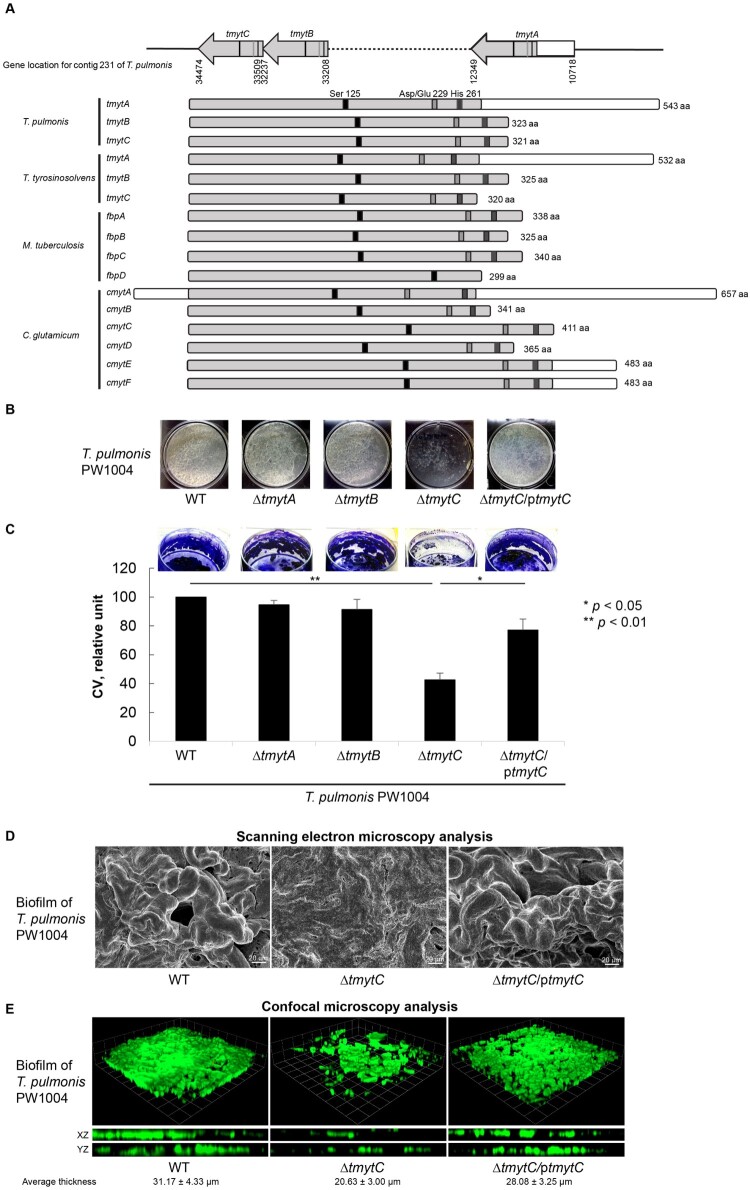
Figure 3.
Characterization of the 3 tmyt homologues in Tsukamurella. (a) Locations of the tmytA, tmytB, and tmytC gene in the genome of T. pulmonis-PW1004 are indicated. Alignment of the tmyt homologues identified in T. pulmonis-PW1004, T. tyrosinosolvens-PW899, M. tuberculosis (GenBank accession numbers NP_218321, NP_216402, YP_177694 and YP_178017) and C. glutamicum (GenBank accession numbers AAAP23202-AAAP232007). The catalytic triad formed by functional residues Ser125, Asp/Glu229, and His261, which are important for mycolyltransferase activity, are indicated by black, grey, and dark grey boxes, respectively. (b) Biofilm formed by the PW1004-WT and its derivative mutants when they were cultured under static conditions for 3 days. With the exception of the PW1004ΔtmytC, dense and confluent biofilm was formed as a floating pellicle at the air-liquid interface in PW1004ΔtmytA, PW1004ΔtmytB, PW1004-WT, and tmytC complemented mutant (PW1004ΔtmytC/ptmytC). (c) Quantitation of the biofilm formed by the PW1004-WT and its derivative mutants using crystal violet staining method. The amount of biofilms was significantly reduced in PW1004ΔtmytC compared to the PW1004-WT (P < 0.01) and complemented mutant (P < 0.05). (d) SEM and (e) confocal microscopy analyses of Tsukamurella biofilms cultured under static conditions for 3 days. Representative SEM micrographs of the biofilm formed by the PW1004ΔtmytC was flattened and less structured compared to those formed by the PW1004-WT and complemented mutant. Likewise, biofilms were fixed and stained with SYTO 9 green fluorescent stain prior to confocal microscopy analysis. Representative micrographs comparing biofilm thickness of each Tsukamurella strain was measured in different points of each field. The means and standard deviations of three independent experiments are shown.