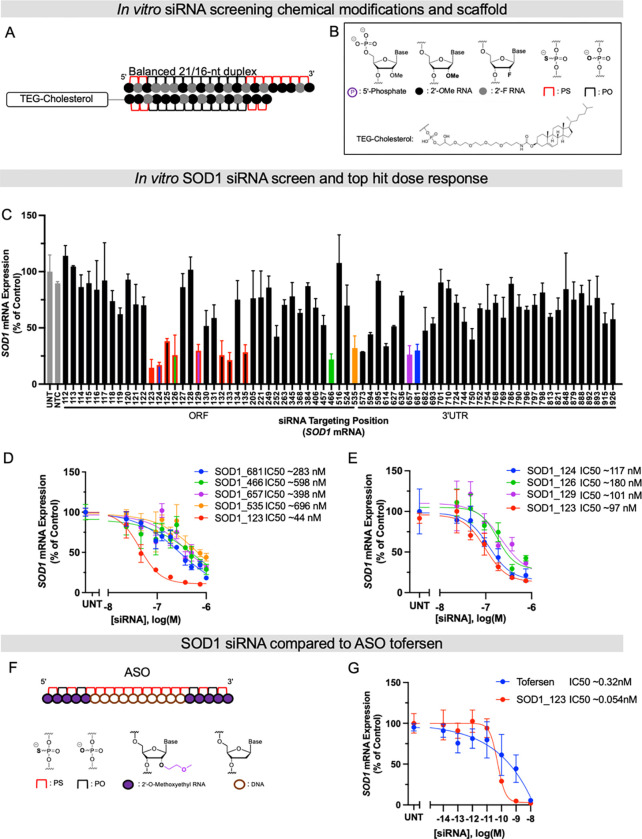
Fig. 1. Systematic screening of chemically modified siRNAs targeting SOD1 mRNA identifies a highly functional compound.
(A and B) Schematics of chemical modifications and siRNA scaffolds tested in screen. (C) In vitro screen of SOD1 siRNAs in HeLa cells. HeLa cells were treated (by passive uptake) with a panel of siRNAs (1.5 μM) for 72 hours. (D) 7-point dose-response curves of lead siRNA compounds identified in primary screen in C for 72 hours. (E) 7-point dose-response curves of lead compounds from siRNA “walk” screen marked in red in the bar graph in C. (F) Schematic of ASO Tofersen and its modifications. (G) 7-point dose-response curves of Tofersen and SOD1_123 with lipid-mediated uptake. All mRNA levels were measured using QuantiGene (n=3, bar is average n=3 ± SD) IC50 values were calculated using the nonlinear least squares method (GraphPad Prism). NTC (nontargeting control siRNA), UNT (untreated).