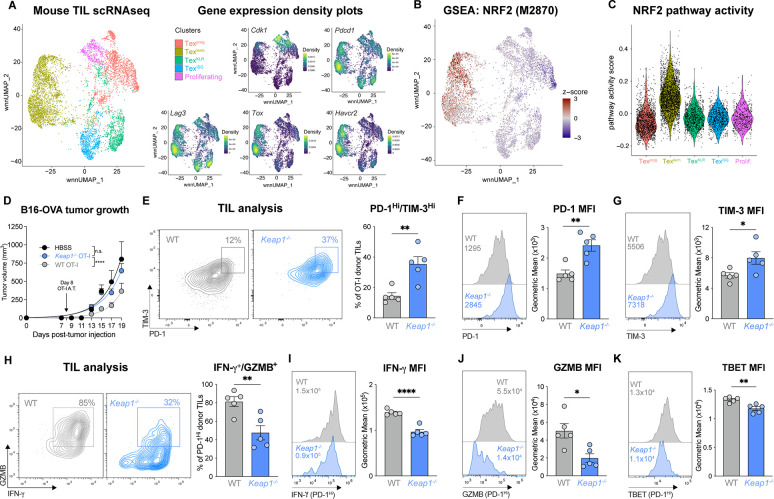
Fig. 2. NRF2 activation is linked to CD8 T cell dysfunction in cancer and poor tumor control.
(A) Weighted nearest neighbor Uniform Manifold Approximation and Projection (wnn_UMAP) of cell-specific transcripts for the 5 major activated (CD44+) CD8+ T cell clusters in B16 tumors (14 days post tumor inoculation (dpti)): progenitor exhausted (Texprog), terminally exhausted (Texterm), exhausted killer cell lectin-like receptor-expressing (TexKLR), exhausted IFN-I stimulated gene expressing (TexISG), and proliferating (prolif.). Gene expression density plots for Cdk1, Pdcd1, Lag3, Tox, and Havcr2 are shown. (B) GSEA embedding of NRF2 transcriptional target signature (M2870) in the CD8 TIL UMAP from (A). (C) Violin plot of NRF2 (M2870) pathway activity score for each of CD8 T cell clusters defined in (A). (D) B16-OVA melanoma tumor growth in mice following adoptive transfer of WT or Keap1−/− OT-I CD8 T cells at 8 dpti (mean±SEM, n=11. (E) PD-1 versus TIM-3 expression on WT and Keap1−/− OT-I CD8 TIL isolated from B16 tumors at 15 dpti (mean±SEM, n=5. (F-G) Histogram of (F) PD-1 and (G) TIM-3 expression on WT or Keap1−/− OT-I CD8 TIL isolated from B16 tumors as in (E), with geometric mean fluorescence intensity (gMFI) shown in inset. Bar graphs quantify the gMFI of PD-1 and TIM-3 expression (mean±SEM, n=5. (H) Representative flow cytometry plot of IFN-γ versus Granzyme B (GZMB) expression for PD-1HI WT and Keap1−/− TIL isolated as in (E). (I-K) Representative histograms and bar graph of gMFI for (I) IFN-γ, (J) GZMB, and (K) TBET expression by PD-1HI WT or Keap1−/− OT-I CD8 TIL isolated from B16 tumors as in (E). *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001.