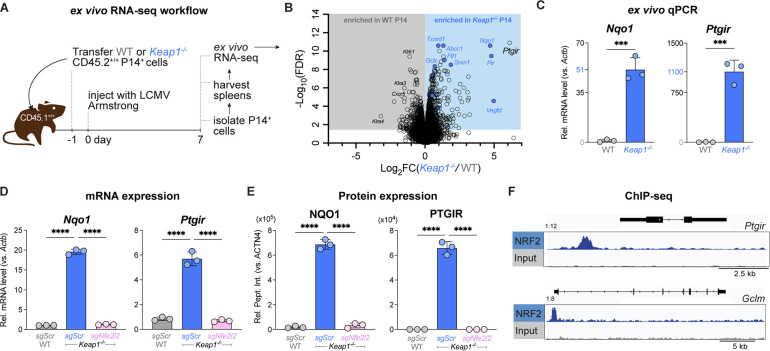
Fig. 4. NRF2 directly regulates expression of the Prostacyclin receptor (Ptgir).
(A) Schematic of RNA-seq workflow for P14 T cells responding to LCMV infection. WT or Keap1−/− P14 T cells (CD45.2+) were adoptively transferred into congenic CD45.1+ hosts followed by infection with LCMV (Armstrong strain). At 7 dpi, P14 T cells were sorted via CD45.2 positive selection for bulk RNA-seq analysis (3 biological replicates/genotype). (B) Volcano plot (horizontal axis: Log2 fold change (Keap1−/−/WT), vertical axis: −Log10 False Discovery Rate (FDR)) highlighting differentially expressed genes (DEGs) in WT and Keap1−/− P14 T cells at 7 dpi. Gray and blue shaded regions represent genes significantly enriched (−Log10 FDR > 1.30, corresponding to FDR <0.05) in WT and Keap1−/− P14 T cells, respectively. Blue circles highlight conventional NRF2 targets from the NFE2L2.V2 gene set (M2870). (C) Relative expression levels of Nqo1 and Ptgir mRNA (normalized to Actb) in WT and Keap1−/− P14 T cells as determined by qPCR (mean±SEM, n=3). (D-E) Relative expression of (D) mRNA and (E) protein levels for NAD(P)H quinone dehydrogenase 1 (Nqo1/NQO1) and Ptgir/PTGIR in activated WT and Keap1−/− P14 cells following CRISPR-Cas9 editing using control (sgScr) or Nfe2l2-targeting (sgNfe2l2) sgRNAs (mean±SEM, n=3). RNA levels were normalized to Actnb. Protein levels were normalized relative to ACTN4 levels. (F) Data tracks for NRF2 peak enrichment at Ptgir (top) and Glcm (bottom) gene loci in CD8 T cells expressing a constitutively active form of NRF2 (CA-NRF2) as determined by NRF2 ChIP-seq. ***P<0.001; ****P<0.0001.