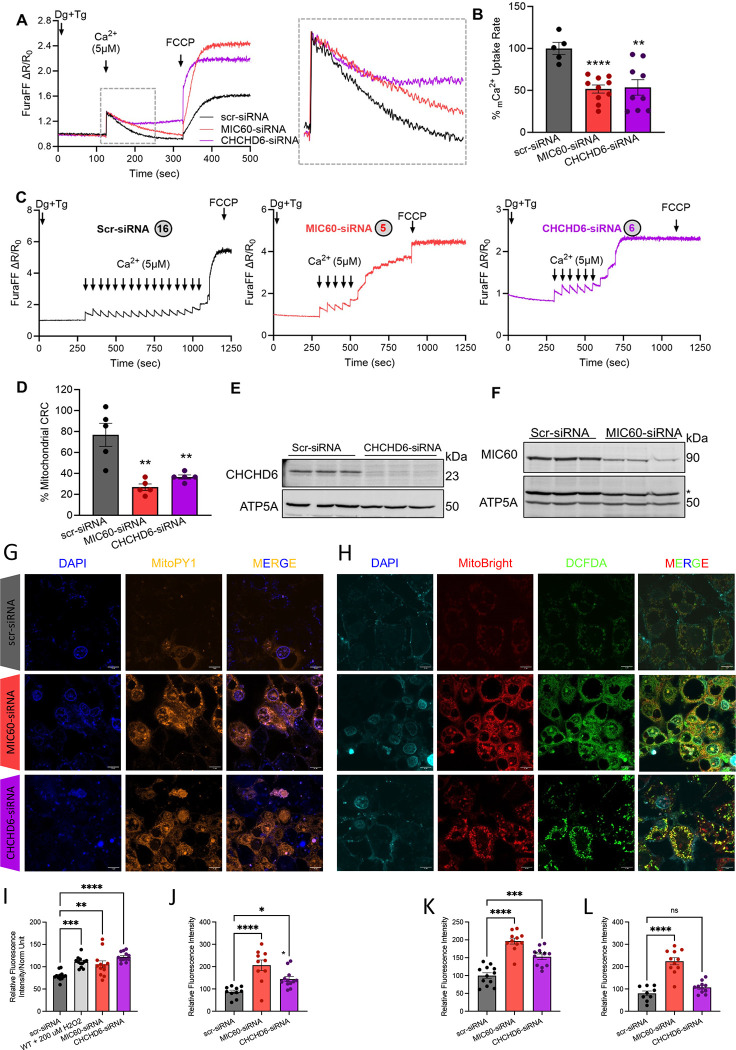
Figure 6: Loss of MIC60 and CHCHD6 in HepG2 cells results in reduced mCa2+ uptake and calcium retention capacity and oxidative stress.
(A) Raw traces showing mitochondrial calcium uptake in permeabilized MIC60 and CHCHD6 knockdown HepG2 cells along with scr-siRNA transected controls. (B) Percentage change in mCa2+ uptake rate quantified from raw traces. (C) Recordings of mitochondrial calcium retention capacity in scr-siRNA, MIC60 siRNA, and CHCHD6 siRNA HepG2 cells. The circles indicate the number of calcium boluses taken up by specific cells. (D) Percentage change in mitochondrial calcium retention capacity quantified from recordings of mitochondrial calcium retention capacity. (E) Immunoblot confirming siRNA-mediated knockdown of CHCHD6 in HepG2 cells. (F) Immunoblot confirming siRNA-mediated knockdown of MIC60 in HepG2 cells. (G) 4′,6-diamidino-2-phenylindole (DAPI) staining, MitoPY1 (5 μM, 45 min at 370 c magnification of 60x), and merge channels in scramble-siRNA (control), MIC60-siRNA, and CHCHD6-siRNA transfected permeabilized HepG2 cells. (H) 4′,6-diamidino-2-phenylindole (DAPI) staining, MitoBright Deep Red (10 μM, 30 min at 37° c), DCFDA (10 μM, 30 min at 37° c, magnification of 60x), and merge channels in scramble-siRNA (control), MIC60-siRNA, and CHCHD6-siRNA transfected permeabilized HEK293 cells. (I) Plate reader-based reactive oxygen species (ROS) quantification. (J) Microscopy-based ROS quantification of MitoPY1 orange, (K) MitoSox Deep Red, and (L) DCFDA. For all statistical tests, one-way ANOVA statistical test was performed with Dunnett’s multiple comparisons test. N=5–10 for all calcium experiments, each indicated by dots, as run in triplicates. N=9–13 for all oxidative stress experiments, each indicated by dots, as run in triplicates. Significance values indicate **P ≤ 0.01 and ****P ≤ 0.0001.