Fig. 2 |. Genomic deletion of UNC13A CE rescues protein levels following TDP-43 knockdown.
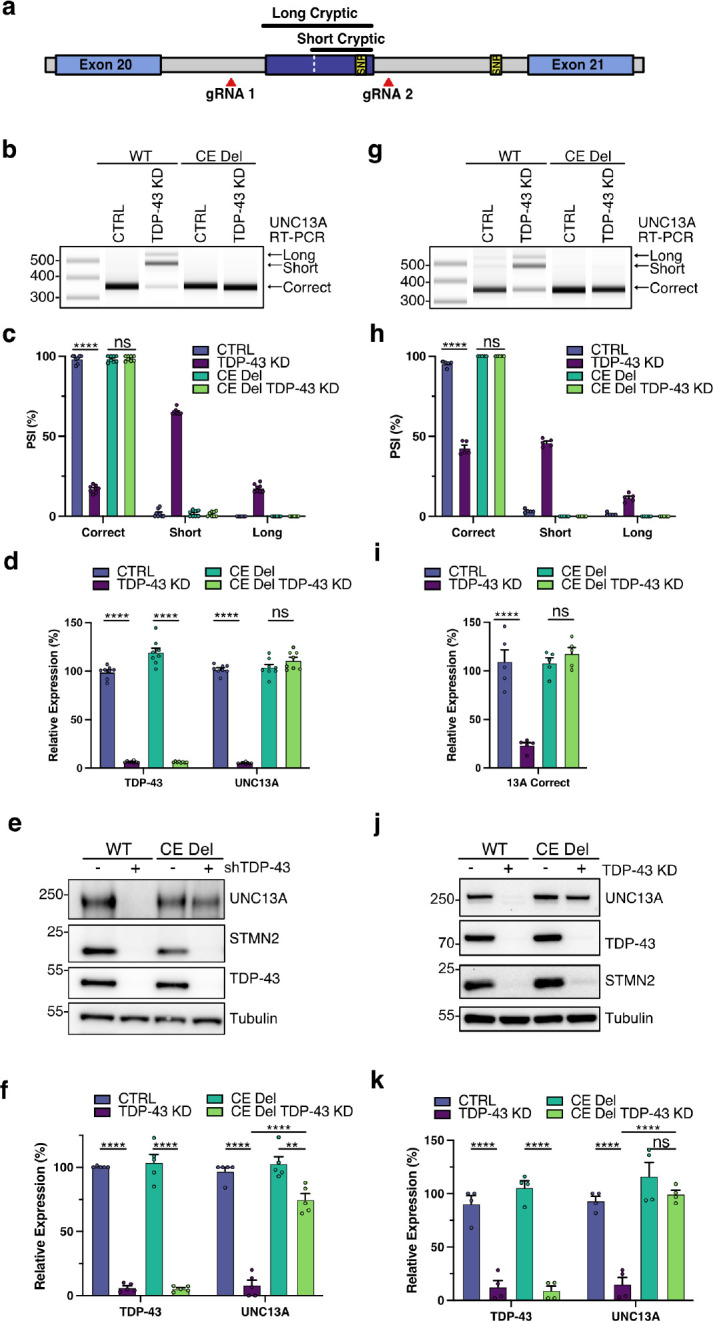
a, Schematic representation of genomic edit of UNC13A cryptic exon. b-f, Analysis of control and TDP-43 KD in WT and CE Del SH-SY5Y cells. (b) RT-PCR analysis of UNC13A splicing indicates CE deletion rescues splicing at the UNC13A exon 20–21 junction after TDP-43 KD. (c) Quantification of results in (B) n=8 biological replicates from 3 experiments. (d) RT-qPCR analysis of TDP-43, correctly spliced UNC13A shows CE deletion rescues UNC13A RNA levels after TDP-43 KD. n = 8 biological replicates from 3 experiments. (e) Western blot analysis of protein lysates shows CE deletion rescues UNC13A protein levels after TDP-43 KD. (f) Quantification of results in (e) n=5 biological replicates from 3 experiments. (g-k) Analysis of control and TDP-43 knockdown in WT and CE Del Halo-TDP iNeurons on DIV 28 following 14 days of HaloProtac treatment. (g) RT-PCR analysis of UNC13A splicing indicates CE deletion rescues splicing at UNC13A exon 20–21 junction after TDP-43 KD. (h) Quantification of results in (g) n=5 biological replicates from 2 experiments. (i) RT-qPCR analysis of correctly spliced UNC13A shows CE deletion rescues UNC13A RNA levels after TDP-43 KD. n=5 biological replicates from 2 experiments. (j) Western blot analysis of protein lysates indicates CE deletion rescues UNC13A protein levels after TDP-43 KD. (k) Quantification of results in (j) n=4 biological replicates from 2 experiments. Graphs for (c) (d) (f) (h) (i) and (k) represent mean ± s.e.m. One-way ANOVA with Tukey multiple comparison test. *P < 0.05; ** P <0.01; *** P <0.001; **** P < 0.0001; ns (not significant).
