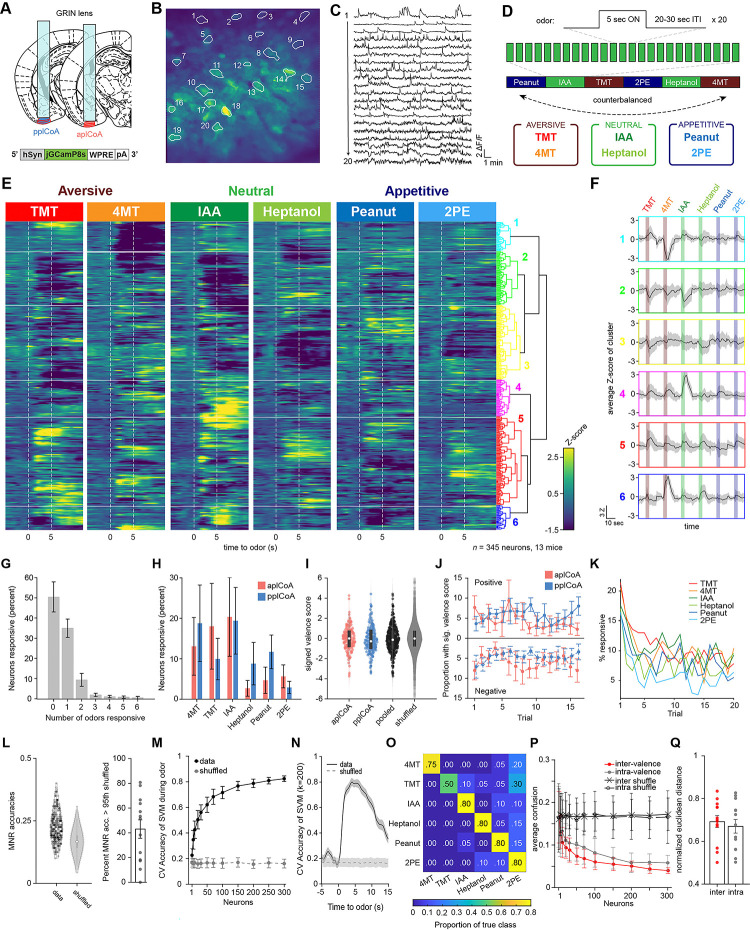
Figure 1. The plCoA encodes innately-valenced odor identity using a population code.
(A) Schematic representation of virus injection and GRIN lens implantation into aplCoA or pplCoA for two-photon microscopy.
(B-C) Representative images (B) and traces (C) of fluorescence changes in individual neurons over an approximately 20-minute period that includes periods of odor stimulation.
(D) Schematic of odor exposure paradigm. Each trial presented 5 seconds of odor followed by a variable inter-trial interval (20–30s). Odors were present in blocks of 20 trials per odor, with 2 counterbalanced block schedules (1 & 2). Six odors were used: the appetitive odors 2-phenylethanol (2PE) and peanut oil (PEA), the neutral odors heptanol (HEP) and isoamyl acetate (IAA), and the aversive odors trimethylthiazoline (TMT) and 4-methylthiazoline (4MT).
(E) Heatmap of trial-averaged and Z-scored odor-evoked activity over time from pooled plCoA neurons. Responses are grouped by hierarchical clustering, with the dendrogram (right). Odor delivery marked by vertical red lines.
(F) Average of trial-averaged and Z-scored odor-evoked activity for each cluster concatenated. The order of color-coded blocks corresponds to the order of clusters in (E).
(G) Proportion of neurons responsive to different numbers of odors. Bars represent the mean across 13 animals and the error bars show SEM.
(H) Proportion responsive to each odor for aplCoA (red) or pplCoA (blue).
(I) Valence scores of individual neurons. White circles show the median of each distribution, whereas the gray rectangle shows the 25th-75th percentile range.
(J) Proportion of neurons with significant valence scores calculated as a function of trial number. Calculated with a 10-trial moving window. Top half shows those with significant positive valence scores, the bottom half shows those with significant negative valence scores.
(K) The percentage of neurons with responses (Z>2 for at least 5 frames) as a function of trial number for each odor.
(L) Left, MNR accuracies for all pooled plCoA neurons (data) and a control distribution where the training labels are shuffled (shuffled) in a violin plot. Right, proportion of neurons in each animal that have MNR accuracy greater than the 95th percentile of the shuffled MNRs.
(M) Cross-validated average accuracies of multinomial SVM’s plotted as a function of the number of neurons used for training during the odor period. Circles represent the mean across 100 iterations of random sampling of neurons and error bars show the standard deviation.
(N) Cross-validated accuracy of ecoc-SVM classifiers for a 6-odor classification task trained using 200 neurons as a function of time. Lines indicate means and shaded areas show the standard deviation across 100 random samplings of 200 neurons from the pooled data, and shuffled training controls where the label vectors are randomly shuffled.
(O) An example confusion matrix for a multinomial SVM trained with 200 neurons.
(Q) Comparison of inter-valence and intra-valence confusion across number of neurons used in training the classifiers. Filled circles show the average of the data across 100 iterations, open circles show shuffled controls.
(4) The normalized average distance between odor pairs that have different valence (inter) or same valence (intra).
Across panels, ns, not significant. Additional specific details of statistical tests can be found in Supplemental Table 1.