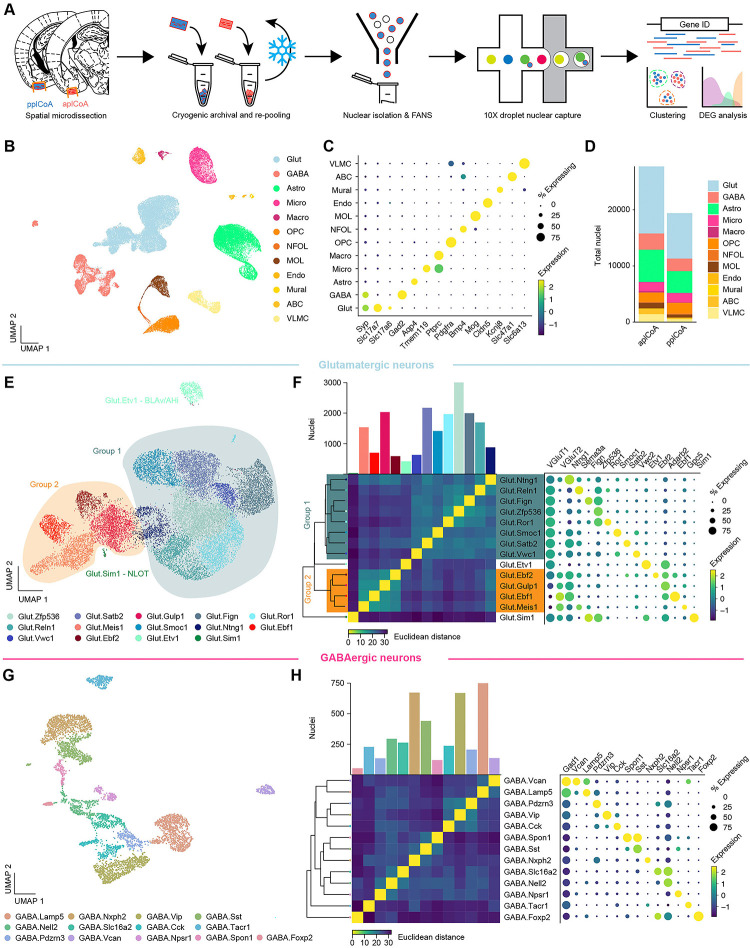
Fig. 3. Transcriptomic heterogeneity of plCoA molecular cell types.
(A) Schematic of freeze and-re-pool strategy for snRNA-seq.
(B) Two-dimensional UMAP (n = 47,132 nuclei, see also Figure S4.3), colored by broad cellular identity assigned by graph-based clustering of neuronal and non-neuronal nuclei.
(C) Cell-type-specific expression of canonical marker genes indicating broad cellular identity in the brain. Dot size is proportional to percentage of nuclei expressing the marker, with color scale representing normalized expression level.
(D) Total proportion of cells of each identified type in each domain of plCoA.
(E) Two-dimensional UMAP of glutamatergic neurons, colored by molecular cell type.
(F) Clustered heatmap showing Euclidean distance between averages of each subtype positioned based on hierarchical clustering (left), and dot plot of marker genes for all glutamatergic subtypes (right).
(G) Two-dimensional UMAP of GABAergic neurons, colored by molecular cell type, like in (E).
(H) Clustered heatmap showing Euclidean distance between averages of each subtype positioned based on hierarchical clustering (left), and dot plot of marker genes for all GABAergic subtypes (right), like in (F).