Fig. 6. Pioneer transcription factor mRNA transfer is indispensable for naïve conversion of hiPSCs.
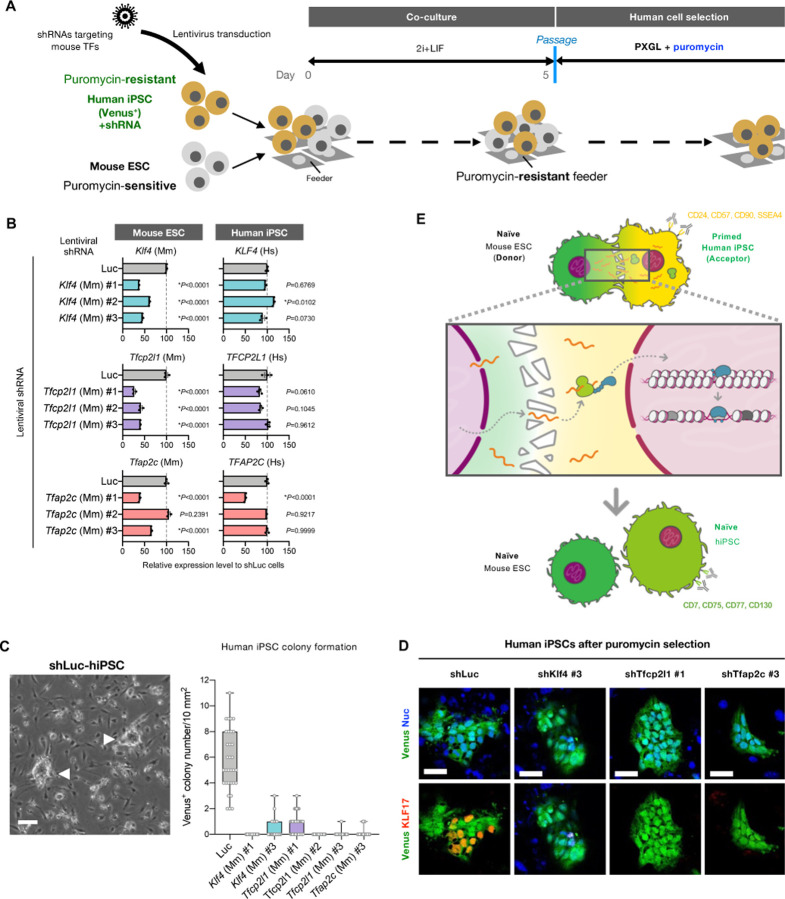
(A) Protocols for coculture of primed hiPSCs expressing Venus and shRNA specifically targeting each mouse transcription factor (puromycin-resistant) with naïve mESCs (puromycin-sensitive) followed by the induction of naïve-like conversion. After the coculture of both cell types (50% each) in 2i+LIF condition for 5 days, they were passed onto the puromycin-resistant feeder cells in the presence of puromycin in PXGL medium to eliminate mESCs. After the puromycin selection, the residual human cells were subsequently analyzed.
(B) RT-qPCR analysis of endogenous Klf4, Tfcp2l1, Tfap2c, Nanog, and Pou5f1 gene expression levels in mESCs in which lentiviral shRNAs targeting mouse Klf4, Tfcp2l1, Tfap2c, Nanog, or Pou5f1 designed in fig. S9A were expressed.
(C) RT-qPCR analysis of endogenous KLF4, TFCP2L1, TFAP2C, NANOG, and POU5F1 gene expression levels in hiPSCs in which lentiviral shRNAs targeting mouse Klf4, Tfcp2l1, Tfap2c, Nanog, or Pou5f1 designed in fig. S9A were expressed.
(D) The emerging colonies of dome-shaped, human cells expressing control luciferase shRNA (shLuc) emerging after puromycin selection is shown (white arrowheads in the left image; scale bar, 100 mm). The number of Venus-positive, dome-shaped colonies emerging after the puromycin selection in each shRNA expression condition was shown in the box and whiskers plot (right; bars represent min to max of the counts).
(E) Immunostaining images of human naïve pluripotency marker KLF17 in Venus-positive hiPSCs expressing shRNAs targeting luciferase (shLuc) or mouse Klf4, Tfcp2l1, and Tfap2c after the coculture with mESCs followed by puromycin selection. Scale bar, 50 mm.
(F) Schematic representation of mESC co-culture-driven reprogramming of primed hPSCs into naïve state via mESC-derived transfer. During the coculture, mRNAs that plays pleiotropic roles including stress responses are transferred between the two cells in a manner dependent on the direct cell contact.