Fig. 1. Datasets characterization and analysis pipeline.
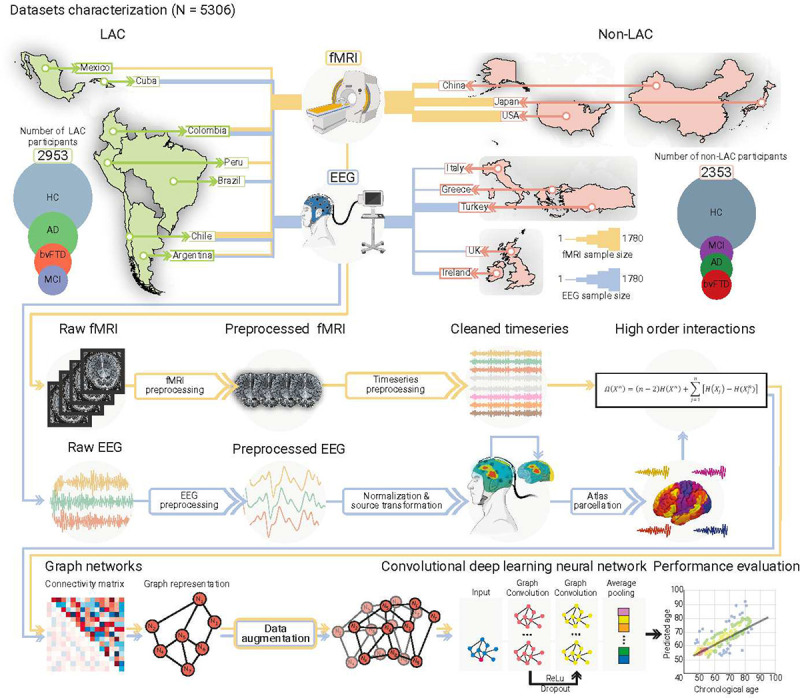
Datasets included Latin American countries (LAC) and non-LAC healthy controls (HC, total N = 3509) and participants with Alzheimer’s disease (AD, total N = 828), behavioral variant frontotemporal dementia (bvFTD, total N = 463), and mild cognitive impairment (MCI, total N = 517). The functional magnetic resonance imaging dataset (fMRI, yellow lines) included 2953 participants from LAC (Argentina, Chile, Colombia, Mexico, and Peru) as well as non-LAC (the USA, China, and Japan). The electroencephalography dataset (EEG, blue lines) involved 2353 participants from Argentina, Brazil, Chile, Colombia, and Cuba (LAC) as well as Greece, Ireland, Italy, Turkey, and the UK (non-LAC). Circles represent the number of participants per group, scaled between the number of participants in the largest and smallest groups for each region to facilitate visualization. Line thickness represents the number of participants with fMRI (yellow lines) and EEG (blue lines) per country. The raw fMRI and EEG signals were preprocessed by filtering and artifact removal and the EEG signals were normalized to project them into source space. A parcellation using the automated anatomical labeling (AAL) atlas for both the fMRI and EEG signals was performed to build the nodes from which we calculated the high-order interactions using the Ω-information metric. A connectivity matrix was obtained for both modalities, which was later represented by graphs. Data augmentation was performed only in the testing dataset. The graphs were used as input for a graph convolutional deep learning network (architecture shown in the last row), with separate models for EEG and fMRI. Finally, age prediction was obtained, and the performance was measured by comparing the predicted vs. the chronological ages. This figure was partially created using Biorender under Team license.
