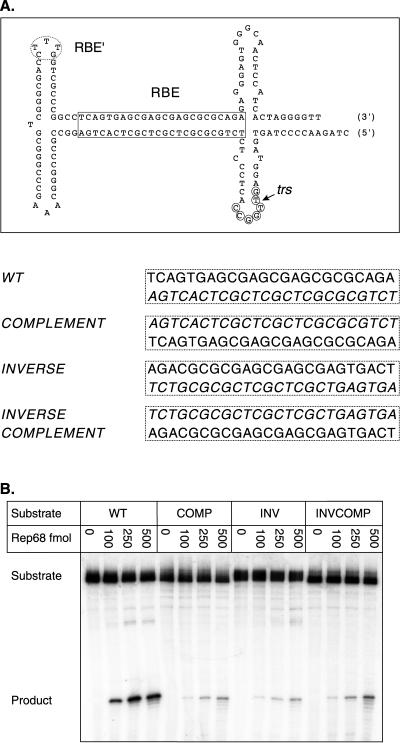
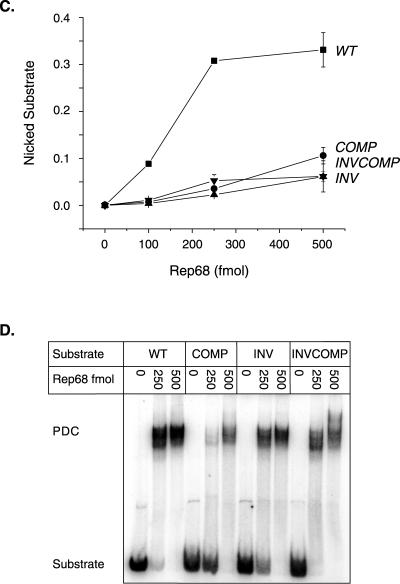
FIG. 4.
Rep endonuclease activity on RBE polarity mutants. (A) The wt AAV TR is depicted after extrusion of the trs stem-loop structure. The RBE is indicated with a box, the RBE′ is indicated with a dashed oval, the minimal trs is indicated with small circles, and the actual nicking site is indicated with a small arrow. The wt RBE was replaced with alternative orientations of this sequence. The sequences of the various RBE orientations are given next to the mutant identifier. Note that the integrity of the RBE base composition is maintained. Only the polarity of the nucleic acid sequence has been altered. (B) Rep68 endonuclease reactions were performed on wt and insertion substrates in the presence of 0.5 mM ATP as described in Materials and Methods. Products were resolved on a 10% denaturing polyacrylamide gel. A representative gel is shown. Numbers above lanes indicate the total amount of Rep68 in the reactions expressed in femtomoles. The positions of substrates and products are indicated. (C) The gels from two independent trials were phosphorimaged, and the amounts of substrate and product were determined. The fractions of nicked substrate for wt and mutant TRs were calculated at each Rep concentration, averaged between trials, and plotted. Closed squares, wt; closed circles, complement; closed triangles, inverse; closed inverted triangles, inverse complement (n = 2 for all data points). Bars indicate the range at each data point in the two independent trials. (D) Rep binding to wt and mutant TRs was assayed under endonuclease conditions in the absence of ATP (see Materials and Methods). Reactions were resolved on a 4% native polyacrylamide gel to separate substrate from protein-bound DNA complexes (PDCs). The positions of substrate and protein-bound DNA complexes are indicated.