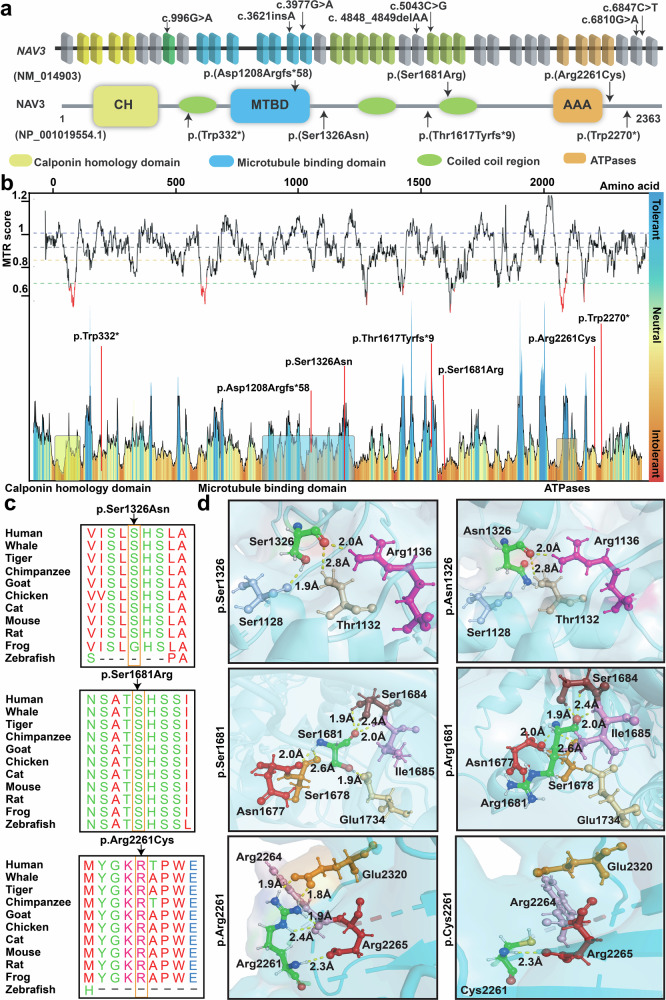
Fig. 2. In-silico analysis and 3D protein modeling support deleterious impact of NAV3 variants.
a Schematic representation of human NAV3 gene (top) and encoded protein (bottom) structures, along with the variants identified in this study. NAV3 in humans is comprised of 39 coding exons, while encodes Neuron Navigator 3 (NAV3) protein with multiple functional domains, including the Calponin homology region (CH), microtubule-binding domain (MTBD), ATPases (AAA), and three coiled-coil regions (green ovals). b The Missense Tolerance Ratio (MTR) graph and Intolerance landscape visualization of NAV3 via MetaDome are presented with relative positions of the variants identified in our cohort. All of the identified variants are located in relatively intolerant regions of the NAV3. c High conservation of the residues, replaced due to the missense variants found in this study, was observed during evolution. d Three-dimensional (3D) protein modeling illustrating an overview of NAV3 protein. The left panels represent wild-type residues at given positions, while their alternate mutated residues are in the right panels. Color code: green element; targeted amino acid; interacting amino acids: mustard, pink, purple, red, badge, rose, and burgundy, while hydrogen bonding between the residues is shown with yellow dotted lines along with the distances in Å. For p.Ser1326Asn substitution, a loss of hydrogen bond with p.Ser1128 (purple) was predicted. Similarly, for the p.Ser1681Arg variant, loss of hydrogen bond with p.Glu1734 (badge) was predicted. The p.Arg2261Cys variant is predicted to abolish four hydrogen bonds: two with p.Arg2264 (pink) and two with p.Glu2320 (mustard).