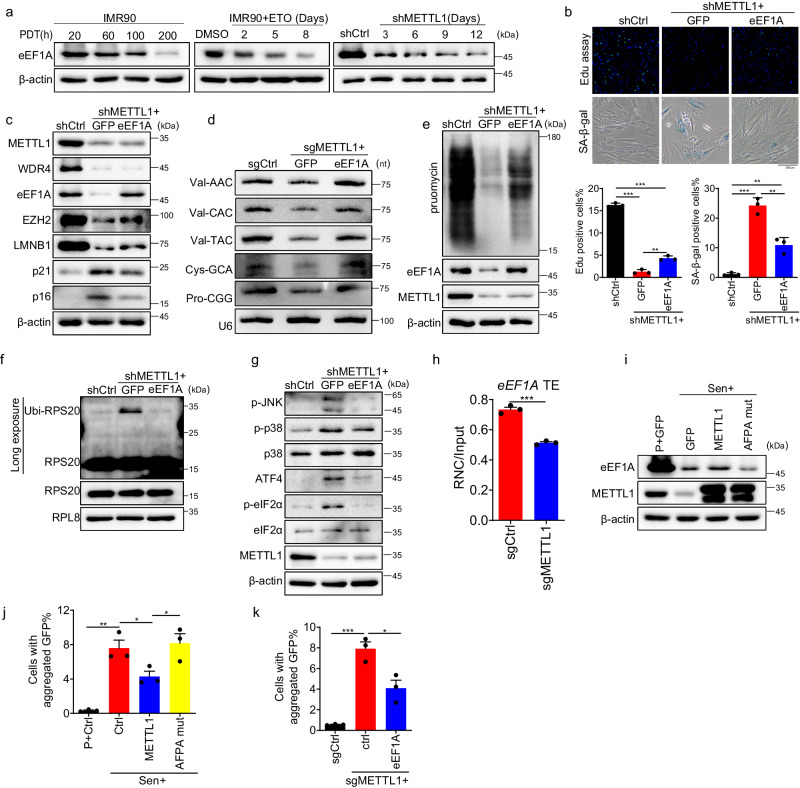
Fig. 7. RTD is involved in the senescence caused by METTL1 deficiency.
a The eEF1A protein level was detected by Western blotting using samples from replicative senescence, ETO-induced senescence, and METTL1-depleted IMR90 cells at various time points as indicated. b Cell proliferation and SA-β-Gal-positive senescent cells were analyzed following eEF1A expression in METTL1-depleted cells. c Senescence markers were analyzed following eEF1A expression in METTL1-depleted cells. d Northern blotting was conducted using METTL1 knockout cells with or without eEF1A overexpression to detect several tRNA level. e Puromycin incorporation was analyzed by Western blotting when eEF1A was expressed in METTL1-depleted cells. f Ubiquitination of RPS20 was assessed in isolated ribosomes from control cells, METTL1 knockout cells, with or without eEF1A overexpression, using a 1 M sucrose cushion, and detected by western blotting.Arrow-pointed bands were ubiquitinated RPS20 protein. g Phosphorylated eIF2α, JNK, p38, and ATF4 proteins were detected by Western blotting in METTL1-depleted cells with or without eEF1A overexpression. h The active translation ratios of eEF1A in METTL1 KO and control cells were determined by qPCR using RNC-mRNA. i eEF1A level was detected in senescent cells overexpressing GFP control, METTL1 and AFPA mutant by Western blotting. The protein aggregation in senescent IMR90 cells with METTL1, AFPA-mutant, or an empty vector control(j) and METTL1 KO cells with or without eEF1a (k) was evaluated using the FlucDM-GFP reporter. The aggregated protein percentage is determined by the aggregated GFP inclusions cell against the total GFP-positive cells. The representative result was shown, three repeats demonstrate similar results (a, c–g, i). All the assays were biologically repeated three times. All data were presented as the mean ± SEM. Two-tailed unpaired t test (h), one-way ANOVA with Bonferroni’s multiple comparisons test (b, j, k). *p < 0.05, **p < 0.01, ***p < 0.001. Source data are provided as a Source Data file.