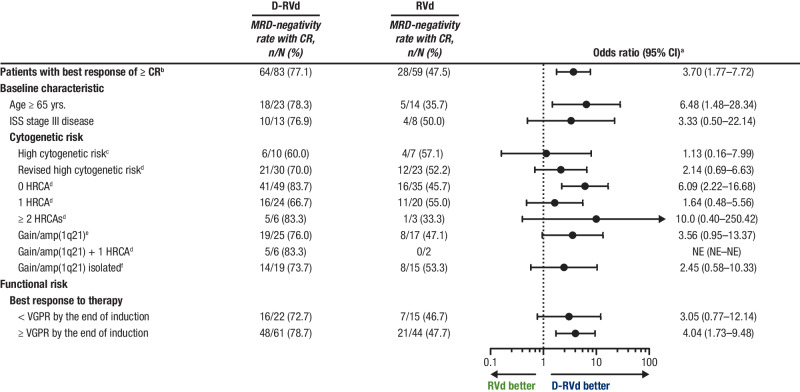
Fig. 2. Subgroup analysis of MRD-negativity (10−5) rates among patients with a best response of ≥ CR by the end of the study.
MRD-negativity rates were evaluated among response-evaluable patientsa who achieved a best response of ≥ CR and were measured at the time of the final analysis (median follow-up in overall population, 49.6 months). MRD was evaluated by next-generation sequencing using the clonoSEQ assay (v2.0; Adaptive Biotechnologies, Seattle, WA) at a minimum sensitivity threshold of 1 in 100,000 cells (10−5) in alignment with IMWG criteria [44]. MRD minimal residual disease, D-RVd daratumumab plus lenalidomide/bortezomib/dexamethasone, RVd lenalidomide/bortezomib/dexamethasone, CI confidence interval, ITT intent-to-treat, ISS International Staging System, HRCA high-risk cytogenetic abnormality, NE not evaluable, VGPR very good partial response, FISH fluorescence in situ hybridization. aMantel–Haenszel estimate of the common odds ratio for unstratified tables is used. An odds ratio > 1 indicates an advantage for D-RVd. bThis analysis included patients from the response-evaluable population, which included all randomized patients who had measurable disease (confirmed MM diagnosis), received ≥ 1 dose of study treatment, and had ≥ 1 postbaseline disease assessment. cHigh-risk cytogenetics are defined based on FISH testing as ≥ 1 of the following: del(17p), t(4;14), or t(14;16). dRevised high-risk cytogenetics are defined based on FISH testing as ≥ 1 HRCA: del(17p), t(4;14), t(14;16), t(14;20), or gain/amp(1q21) (≥ 3 copies of chromosome 1q21). ePatients in this group have gain/amp(1q21) with or without other HRCAs (del[17p], t[4;14], t[14;16], or t[14;20]). fPatients with isolated gain/amp(1q21) do not have any other HRCAs.